Abstract
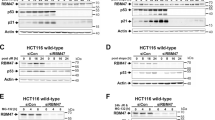
We identified IFI16 as a BRCA1-associated protein involved in p53-mediated apoptosis. IFI16 contains the Pyrin/PAAD/DAPIN domain, commonly found in cell death-associated proteins. BRCA1 (aa 502–802) interacted with the IFI16 Pyrin domain (aa 1–130). We found that IFI16 was localized in the nucleoplasm and nucleoli. Clear nucleolar IFI16 localization was not observed in HCC1937 BRCA1 mutant cells, but reintroduction of wild-type BRCA1 restored IFI16 nuclear relocalization following IR (ionizing radiation). Coexpression of IFI16 and BRCA1 enhanced DNA damage-induced apoptosis in mouse embryonic fibroblasts from BRCA1 mutant mice expressing wild-type p53, although mutant IFI16 deficient in binding to BRCA1 did not induce apoptosis. Furthermore, tetracycline-induced IFI16 collaborated in inducing apoptosis when adenovirus p53 was expressed in DNA-damaged p53-deficient EJ cells. These results indicate a BRCA1-IFI16 role in p53-mediated transmission of DNA damage signals and apoptosis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Abbreviations
- BASC:
-
BRCA1-associated genome surveillance complex
- MEF:
-
mouse embryonic fibroblast
- HEK293:
-
human embryonic kidney 293
- IR:
-
ionizing radiation
References
Alberg AJ and Helzlsouer KJ . (1997). Curr. Opin. Oncol., 9, 505–511.
Aravind L, Dixit VM and Koonin EV . (2001). Science, 291, 1279–1284.
Bork P, Hofmann K, Bucher P, Neuwald AF, Altschul SF and Koonin EV . (1997). FASEB J., 11, 68–76.
Cortez D, Wang Y, Qin J and Elledge SJ . (1999). Science, 286, 1162–1166.
DeYoung KL, Ray ME, Su YA, Anzick SL, Johnstone RW, Trapani JA, Meltzer PS and Trent JM . (1997). Oncogene, 15, 453–457.
Fairbrother WJ, Gordon NC, Humke EW, O'Rourke KM, Starovasnik MA, Yin JP and Dixit VM . (2001). Protein Sci., 10, 1911–1918.
Gu W, Malik S, Ito M, Yuan C-X, Fondell JD, Zhang X, Martinez E, Qin J and Roeder RG . (1999). Mol. Cell., 3, 97–108.
Harkin DP, Bean JM, Miklos D, Song YH, Truong VB, Englert C, Christians FC, Ellisen LW, Maheswaran S, Oliner JD and Haber DA . (1999). Cell, 97, 575–586.
Johnstone RW, Kerry JA and Trapani JA . (1998). J. Biol. Chem., 273, 17172–17177.
Johnstone RW and Trapani JA . (1999). Mol. Cell. Biol., 19, 5833–5838.
Johnstone RW, Wei W, Greenway A and Trapani JA . (2000). Oncogene, 19, 6033–6042.
Kaelin WG Jr, Pallas DC, DeCaprio JA, Kaye FJ and Livingston DM . (1991). Cell, 64, 521–532.
Koonin VF, Altschul SF and Bork P . (1996). Nat. Genet., 13, 266–267.
Lee SW, Fang L, Igarashi M, Ouchi T, Lu K-P and Aaronson SA . (2000). Proc. Natl. Acad. Sci. USA, 97, 8302–8305.
Martinon F, Hofmanndouble K and Tschopp J . (2001). Curr. Biol., 11, 118–120.
Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA and Harshman K et al. (1994). Science, 266, 66–71.
Nathanson KN, Wooster R and Weber BL . (2001). Nat. Med., 7, 552–556.
Okada S and Ouchi T . (2003). J. Biol. Chem., 278, 2015–2020.
Ouchi T, Monteiro AN, August A, Aaronson SA and Hanafusa H . (1998). Proc. Natl. Acad. Sci. USA, 95, 2302–2306.
Ouchi T, Lee SW, Ouchi M, Aaronson SA and Horvath CM . (2000). Proc. Natl. Acad. Sci. USA, 97, 5208–5213.
Pawlowski K, Pio F, Chu Z, Reed JC and Godzik A . (2001). Trends Biochem. Sci., 26, 85–87.
Pear WS, Nolan GP, Scott ML and Baltimore D . (1993). Proc. Natl. Acad. Sci. USA, 990, 8392–8396.
Scully R, Chen J, Plug A, Xiao Y, Weaver D, Feunteun J, Ashley T and Livingston DM . (1997a). Cell, 88, 265–275.
Scully R, Chen J, Ochs RL, Keegan K, Hoekstra M, Feunteun J and Livingston DM . (1997b). Cell, 90, 425–435.
Scully R, Ganesan S, Vlasakova K, Chen J, Socolovsky M and Livingston DM . (1999). Mol. Cell, 4, 1093–1099.
Somasundaram K, MacLachlan TK, Burns TF, Sgagias M, Cowan KH, Weber BL and El-Deiry WS . (1999). Oncogene, 18, 6605–6614.
Staub E, Dahl E and Rosenthal A . (2001). Trends Biochem. Sci., 26, 83–85.
Tomlinson GE, Chen TT, Stastny VA, Virmani AK, Spillman MA, Tonk V, Blum JL, Schneider NR, Wistuba II, Shay JW, Minna JD and Gazdar AF . (1998). Cancer Res., 58, 3237–3242.
Trapani JA, Browne KA, Dawson MJ, Ramsay RG, Eddy RL, Show TB, White PC and Dupont B . (1992). Immunogenetics, 36, 369–376.
Wang Y, Cortez D, Yazdi P, Neff N, Elledge SJ and Qin J . (2000). Genes Dev., 14, 927–939.
Xu X, Wagner KU, Larson D, Weaver Z, Li C, Ried T, Hennighausen L, Wynshaw-Boris A and Deng C-X . (1999). Nat. Genet., 22, 37–43.
Xu X, Qiao W, Linke SP, Cao L, Li WM, Furth PA, Harris CC and Deng C-X . (2001). Nat. Genet., 28, 266–271.
Xu B, O'Donnell AH, Kim S-T and Kastan MB . (2002). Cancer Res., 62, 4588–4591.
Zhang H, Somasundaram K, Peng Y, Tian H, Zhang H, Bi D, Weber BL and El-Deiry WS . (1998). Oncogene, 16, 1713–1721.
Zheng L, Pan H, Li S, Flesken-Nikitin A, Chen P-L, Boyer T-G and Lee W-H . (2000). Mol. Cell, 6, 757–768.
Acknowledgements
We are grateful to Drs David M Livingston and George R Stark for helpful discussion, Ralph Scully for HCC1937-BRCA1 cells, Mutsuko Ouchi for generating and characterizing anti-BRCA1 monoclonal antibodies, Dr Scott Henderson, Jane Sue and Bryan Kloos for confocal laser microscope analysis, and the Mount Sinai School of Medicine Core Facilities for the monoclonal antibody, DNA sequencing and microscopic analysis. This work was supported by an NIH Predoctoral Training Award (JAA), the New York City Council Speaker's Fund (TO), an EMPIRE Grant of State of New York (TO) and National Cancer Institute Awards CA80058, CA78356 (SWL), CA84199 (JQ) and CA79892 and CA90631 (TO).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Aglipay, J., Lee, S., Okada, S. et al. A member of the Pyrin family, IFI16, is a novel BRCA1-associated protein involved in the p53-mediated apoptosis pathway. Oncogene 22, 8931–8938 (2003). https://doi.org/10.1038/sj.onc.1207057
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.onc.1207057
Keywords
This article is cited by
-
Interferon-Gamma-Inducible Protein 16 Inhibits Hepatocellular Carcinoma via Interferon Regulatory Factor 3 on Chemosensitivity
Digestive Diseases and Sciences (2024)
-
A Bayesian network structure learning approach to identify genes associated with stress in spleens of chickens
Scientific Reports (2022)
-
Comprehensive analysis identifies IFI16 as a novel signature associated with overall survival and immune infiltration of skin cutaneous melanoma
Cancer Cell International (2021)
-
Cytosolic DNA Sensor Upregulation Accompanies DNA Electrotransfer in B16.F10 Melanoma Cells
Molecular Therapy - Nucleic Acids (2016)
-
Nutlin-3-induced redistribution of chromatin-bound IFI16 in human hepatocellular carcinoma cells in vitro is associated with p53 activation
Acta Pharmacologica Sinica (2015)