Abstract
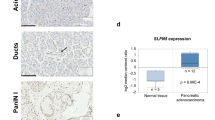
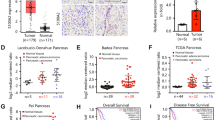
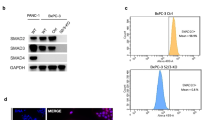
Recent data suggest that SEL1L may play an important role in pancreatic carcinoma, similar to breast cancer, where the expression of SEL1L has been associated with a reduction in both proliferative activity in vitro and clinical tumor aggressiveness. To investigate this possibility, we examined the expression of Sel1L in a series of primary pancreatic carcinomas by immunohistochemistry and characterized the effects of Sel1L overexpression both in vitro and in vivo. In 74 pancreatic cancers analysed, 36% lacked Sel1L expression, although there was no significant correlation between the expression of Sel1L and any clinicopathologic parameter, including survival. However, immunohistochemical reactivity for Sel1L and Dpc4/Smad4 was concordant in 69% of cases (χ2 test P<0.004). Overexpression of SEL1L in stably transfected pancreatic cancer cells caused both a decrease in clonogenicity and anchorage-independent growth as well as a significant increase in the levels of activin A and SMAD4. When implanted in nude mice, Suit-2-SEL1L-overexpressing clones displayed a considerably reduced rate of tumor growth. Thus, it can be hypothesized that Sel1L plays an important function in the growth and aggressiveness of pancreatic carcinoma. Moreover, our data provide evidence that SEL1L has an impact on the expression of genes involved in regulation of cellular growth, possibly through the TGF-β signaling pathway.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Abbreviations
- Mab:
-
monoclonal antibody
- DEX:
-
dexamethasone
- RT–PCR:
-
reverse transcription–PCR
- HPRT:
-
hypoxanthine phosphoribosyltransferase
References
Apelqvist A, Li H, Sommer L, Beatus P, Anderson DJ, Honjo T, Hrabe de Angelis M, Lendahl U and Edlund H . (1999). Nature, 400, 877–881.
Artavanis-Tsakonas S, Rand MD and Lake RJ . (1999). Science, 284, 770–776.
Biunno I, Appierto V, Cattaneo M, Leone BE, Balzano G, Socci C, Saccone S, Letizia A, Della Valle G and Sgaramella V . (1997). Genomics, 46, 284–286.
Biunno I, Bernard L, Dear P, Cattaneo M, Volorio S, Zannini L, Bankier A and Zollo M . (2000). Hum. Genet., 106, 227–235.
Biunno I, Castiglioni B, Rogozin IB, DeBellis G, Malferrari G and Cattaneo M . (2002). Omics., 6, 187–198.
Capobianco AJ, Zagouras P, Blaumueller CM, Artavanis-Tsakonas S and Bishop JM . (1997). Mol. Cell. Biol., 17, 6265–6273.
Cattaneo M, Orlandi R, Ronchini C, Granelli P, Malferrari G, Menard S and Biunno I . (2000). Int. J. Biol. Markers, 15, 26–32.
Cattaneo M, Sorio C, Malferrari G, Rogozin IB, Bernard L, Scarpa A, Zollo M and Biunno I . (2001). DNA Cell. Biol., 20, 1–9.
Chen YG, Lui HM, Lin SL, Lee JM and Ying SY . (2002). Exp. Biol. Med. (Maywood), 227, 75–87.
Cocolakis E, Lemay S, Ali S and Lebrun JJ . (2001). J. Biol. Chem., 276, 18430–18436.
Cook PW, Swanson KT, Edwards CP and Firestone GL . (1988). Mol. Cell. Biol., 8, 1449–1459.
Donoviel DB, Donoviel MS, Fan E, Hadjantonakis A and Bernstein A . (1998). Mech. Dev., 78, 203–207.
Ellisen LW, Bird J, West DC, Soreng AL, Reynolds TC, Smith SD and Sklar J . (1991). Cell., 66, 649–661.
Furue M, Zhang Y, Okamoto T, Hata RI and Asashima M . (2001). Biochem. Biophys. Res. Commun., 282, 745–749.
Grant B and Greenwald I . (1996). Genetics, 143, 237–247.
Grant B and Greenwald I . (1997). Development, 124, 637–644.
Greenwald I . (1998). Genes. Dev., 12, 1751–1762.
Hampton RY, Gardner RG and Rine J . (1996). Mol. Biol. Cell., 7, 2029–2044.
Harada Y, Ozaki K, Suzuki M, Fujiwara T, Takahashi E, Nakamura Y and Tanigami A . (1999). J. Hum. Genet., 44, 330–336.
Jensen J, Heller RS, Funder-Nielsen T, Pedersen EE, Lindsell C, Weinmaster G, Madsen OD and Serup P . (2000). Diabetes, 49, 163–176.
Jhappan C, Gallahan D, Stahle C, Chu E, Smith GH, Merlino G and Callahan R . (1992). Genes. Dev., 6, 345–355.
Kløppel G, Hruban R, Longnecker D, Adler G, Kern S and Partanen T . (2000). Pathology and Genetics of Tumours of the Digestive System: WHO lassification of Tumours. Hamilton S, Aaltonen L (eds). IARC press: Lyon, pp 219–250.
Kudawara I, Ueda T, Yoshikawa H, Miyama T, Yamamoto T and Nishizawa Y . (2001). Eur. J. Cancer, 37, 1703–1708.
Lei J, Zou TT, Shi YQ, Zhou X, Smolinski KN, Yin J, Souza RF, Appel R, Wang S, Cymes K, Chan O, Abraham JM, Harpaz N and Meltzer SJ . (1996). Oncogene, 13, 2459–2462.
Liu QY, Niranjan B, Gomes P, Gomm JJ, Davies D, Coombes RC and Buluwela L . (1996). Cancer Res., 56, 1155–1163.
Massague J, Blain SW and Lo RS . (2000). Cell, 103, 295–309.
Moore P, Orlandini S, Zamboni G, Capelli P, Rigaud G, Falconi M, Bassi C, Lemoine N and Scarpa A . (2001a). Br. J. Cancer, 84, 253–262.
Moore P, Sipos B, Orlandini S, Sorio C, Real F, Lemoine N, Gress T, Bassi C, Klöppel G, Kalthoff H, Löhr M and Scarpa A . (2001b). Virch. Arch., 439, 798–802.
Norman J, Franz M, Schiro R, Nicosia S, Docs J, Fabri PJ and Gower Jr WR . (1994). J. Surg. Res., 57, 33–38.
Orlandi R, Cattaneo M, Troglio F, Campiglio M, Biunno I and Menard S . (2002a). Int. J. Biol. Markers, 17, 104–111.
Orlandi R, Cattaneo M, Troglio F, Casalini P, Ronchini C, Menard S and Biunno I . (2002b). Cancer Res., 62, 567–574.
Robbins J, Blondel BJ, Gallahan D and Callahan R . (1992). J. Virol., 66, 2594–2599.
Roberts AB . (1999). Microbes Infect, 1, 1265–1273.
Rooke HM and Crosier KE . (2001). Pathology, 33, 73–84.
Rozenblum E, Schutte M, Goggins M, Hahn SA, Panzer S, Zahurak M, Goodman SN, Sohn TA, Hruban RH, Yeo CJ and Kern SE . (1997). Cancer Res., 57, 1731–1734.
Scarpa A, Orlandini S, Moore P, Lemoine N, Beghelli S, Baron A, Falconi M and Zamboni G . (2002). Virch. Arch., 440, 155–159.
Schutte M, Hruban RH, Hedrick L, Cho KR, Nadasdy GM, Weinstein CL, Bova GS, Isaacs WB, Cairns P, Nawroz H, Sidransky D, Casero Jr RA, Meltzer PS, Hahn SA and Kern SE . (1996). Cancer Res., 56, 2527–2530.
Schwarte-Waldhoff I, Klein S, Blass-Kampmann S, Hintelmann A, Eilert C, Dreschers S, Kalthoff H, Hahn SA and Schmiegel W . (1999). Oncogene, 18, 3152–3158.
Schwarte-Waldhoff I, Volpert OV, Bouck NP, Sipos B, Hahn SA, Klein-Scory S, Luttges J, Kloppel G, Graeven U, Eilert-Micus C, Hintelmann A and Schmiegel W . (2000). Proc. Natl. Acad. Sci. USA, 97, 9624–9629.
Shelly LL, Fuchs C and Miele L . (1999). J. Cell. Biochem., 73, 164–175.
Tascilar M, Skinner HG, Rosty C, Sohn T, Wilentz RE, Offerhaus GJ, Adsay V, Abrams RA, Cameron JL, Kern SE, Yeo CJ, Hruban RH and Goggins M . (2001). Clin. Cancer Res., 7, 4115–4121.
Thompson Jr EA . (1989). Cancer Res., 49, 2259s–2265s.
Weinmaster G . (1997). Mol. Cell Neurosci., 9, 91–102.
Acknowledgements
This work was supported by grants from Associazione Italiana Ricerca Cancro, Milan, Italy to AS; Ministero Università (Cofin 2002068231 and 2001068593), Rome, Italy; European Community Grant QLG1-CT-2002-01196. MC is supported by a fellowship from Fondazione Italiana Ricerca sul Cancro (FIRC). PSM is supported by Fondazione Cassa di Risparmio di Verona (Bando 2001), Verona, Italy.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Cattaneo, M., Orlandini, S., Beghelli, S. et al. SEL1L expression in pancreatic adenocarcinoma parallels SMAD4 expression and delays tumor growth in vitro and in vivo. Oncogene 22, 6359–6368 (2003). https://doi.org/10.1038/sj.onc.1206665
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.onc.1206665
Keywords
This article is cited by
-
SEL1L, an UPR Response Protein, a Potential Marker of Colonic Cell Transformation
Digestive Diseases and Sciences (2012)
-
Recognition of Host Proteins by Helicobacter Cysteine-Rich Protein C
Current Microbiology (2011)
-
Hypoxia induces p53-dependent transactivation and Fas/CD95-dependent apoptosis
Cell Death & Differentiation (2007)