Abstract
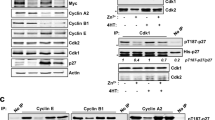
c-fos gene is expressed constitutively in a number of tissues as well as in certain tumor cells and is inducible, in general rapidly and transiently, in virtually all other cell types by a variety of stimuli. Its protein product, c-Fos, is a short-lived transcription factor that heterodimerizes with various protein partners within the AP-1 transcription complex via leucine zipper/leucine zipper interactions for binding to specific DNA sequences. It is mostly, if not exclusively, degraded by the proteasome. To localize the determinant(s) responsible for its instability, we have conducted a genetic analysis in which the half-lives of c-Fos mutants and chimeras made with the stable EGFP reporter protein were compared under two experimental conditions taken as example of continous and inducible expression. Those were constitutive expression in asynchronously growing Balb/C 3T3 mouse embryo fibroblasts and transient induction in the same cells undergoing the G0/G1 phase transition upon stimulation by serum. Our work shows that c-Fos is degraded faster in synchronous- than in asynchronous cells. This difference in turnover is primarily accounted for by several mechanisms. First, in asynchronous cells, a unique C-terminal destabilizer is active whereas, in serum-stimulated cells two destabilizers located at both extremities of the protein are functional. Second, heterodimerization and/or binding to DNA accelerates protein degradation only during the G0/G1 phase transition. Adding another level of complexity to turnover control, phosphorylation at serines 362 and 374, which are c-Fos phosphorylation sites largely modified during the G0/G1 phase transition, stabilizes c-Fos much more efficiently in asynchronous than in serum-stimulated cells. In both cases, the reduced degradation rate is due to inhibition of the activity of the C-terminal destabilizer. However, in serum-stimulated cells, this effect is partially masked by the activation of the N-terminal destabilizer and basic domain/leucine zipper-dependent mechanisms. Taken together, our data show that multiple degradation mechanisms, differing according to the conditions of expression, may operate on c-Fos to ensure a proper level and/or timing of expression. Moreover, they also indicate that the half-life of c-Fos during the G0/G1 phase transition is determined by a delicate balance between opposing stabilizing and destabilizing mechanisms operating at the same time.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Abbreviations
- DBD:
-
DNA-binding domain
- LZ:
-
leucine zipper
- bZIP:
-
basic DNA-binding domain/leucine zipper
- EGFP:
-
Enhanced green fluorescent protein
References
Abate C, Luk D and Curran T . (1990). Cell Growth Diff., 1, 455–462.
Acquaviva C, Brockly F, Ferrara P, Bossis G, Salvat C, Jariel-Encontre I and Piechaczyk M . (2001a). Oncogene, 20, 7563–7572.
Acquaviva C, Salvat C, Brockly F, Bossis G, Ferrara P, Piechaczyk M and Jariel-Encontre I . (2001b). Oncogene, 20, 7563–7572.
Angel P, Szabowski A and Schorpp-Kistner M . (2001). Oncogene, 20, 2413–2423.
Bakiri L, Matsuo K, Wisniewska M, Wagner EF and Yaniv M . (2002). Mol. Cell. Biol., 22, 4952–4964.
Chaussepied M, Lallemand D, Moreau MF, Adamson R, Hall R and Langsley G . (1998). Mol. Biochem. Parasitol., 94, 215–226.
Chen J, Kelz MB, Hope BT, Nakabeppu Y and Nestler EJ . (1997). J. Neurosci., 17, 4933–4941.
Chen RH, Abate C and Blenis J . (1993). Proc. Natl. Acad. Sci. USA, 90, 10952–10956.
Chen RH, Juo PC, Curran T and Blenis J . (1996). Oncogene, 12, 1493–1502.
Chinenov Y and Kerppola TK . (2001). Oncogene, 20, 2438–2452.
Coux O . (2002). Prog. Mol. Subcell Biol., 29, 85–107.
Curran T . (1988). The Oncogene Handbook. Elsevier: Amsterdam, pp 307–325.
DeMartino GN and Slaughter CA . (1999). J. Biol. Chem., 274, 22123–22126.
Dobrazanski P, Noguchi T, Kovary K, Rizzo CA, Lazo PS and Bravo R . (1991). Mol. Cell. Biol., 11, 5470–5478.
Eibl B, Greiter E, Grunewald K, Gastl G, Weyrer K, Thaler J, Aulitzky W, Herrmann F, Rapp U and Huber C . (1995). Cytokines Mol. Ther., 1, 29–38.
Ferdous A, Gonzalez F, Sun L, Kodadek T and Johnston SA . (2001). Mol. Cell, 7, 981–991.
Fujii M, Niki T, Mori T, Matsuda T, Matsui M, Nomura N and Seiki M . (1991). Oncogene, 6, 1023–1029.
Gamberi G, Benassi MS, Bohling T, Ragazzini P, Molendini L, Sollazzo MR, Pompetti F, Merli M, Magagnoli G, Balladelli A and Picci P . (1998). Oncology, 55, 556–563.
Gianni M, Bauer A, Garattini E, Chambon P and Rochette-Egly C . (2002). EMBO J., 21, 3760–3769.
Glickman MH and Ciechanover A . (2002). Physiol. Rev., 82, 373–428.
Gonzalez F, Delahodde A, Kodadek T and Johnston SA . (2002). Science, 296, 548–550.
Gruda MC, Kovary K, Metz R and Bravo R . (1994). Oncogene, 9, 2537–2547.
Hafezi F, Steinbach JP, Marti A, Munz K, Wang ZQ, Wagner EF, Aguzzi A and Reme CE . (1997). Nat. Med., 3, 346–349.
He H, Qi XM, Grossmann J and Distelhorst CW . (1998). J. Biol. Chem., 273, 25015–25019.
Herdegen T and Waetzig V . (2001). Oncogene, 20, 2424–2437.
Hermida-Matsumoto ML, Chock PB, Curran T and Yang DC . (1996). J. Biol. Chem., 271, 4930–4936.
Huang TS, Kuo ML, Lin JK and Hsieh JS . (1995). Cancer Lett., 96, 1–7.
Iwahashi M, Otsuka M, Shimotakahara S, Uozumi K, Hanada S, Arima T and Matsumoto M . (1994). Br. J. Haematol., 86, 193–194.
Jochum W, Passegue E and Wagner EF . (2001). Oncogene, 20, 2401–2412.
Johnson RS, Spiegelman BM and Papaioannou V . (1992). Cell, 71, 577–586.
Kanaitsuka T, Ishii K, Zu Y, Ashihara T, Hanaoka M and Namba Y . (1988). Acta. Pathol. Jpn., 38, 1523–1536.
Kanaitsuka T, Itani K, Shigeta H, Yamamura Y, Kogawa T, Yoshikawa T, Sugino S, Kanatsuna T, Kondou M, Takashina K . (1987). Nippon Naika Gakkai Zasshi, 76, 1595–1603.
Kim YH, Choi MR, Song DK, Huh SO, Jang CG and Suh HW . (2000). Brain Res., 872, 227–230.
Kisselev AF and Goldberg AL . (2001). Chem. Biol., 8, 739–758.
Kovary K and Bravo R . (1991). Mol. Cell. Biol., 11, 2451–2459.
Kovary K and Bravo R . (1992). Mol. Cell. Biol., 12, 5015–5023.
Krappmann D, Wulczyn FG and Scheidereit C . (1996). EMBO J., 15, 6716–6726.
Lallemand D, Spyrou G, Yaniv M and Pfarr CM . (1997). Oncogene, 14, 819–830.
Li X, Zhao X, Fang Y, Jiang X, Duong T, Fan C, Huang CC and Kain SR . (1998). J. Biol. Chem., 273, 34970–34975.
Macian F, Lopez-Rodriguez C and Rao A . (2001). Oncogene, 20, 2476–2489.
Malek NP, Sundberg H, McGrew S, Nakayama K, Kyriakides TR, Roberts JM and Kyriakidis TR . (2001). Nature, 413, 323–327.
Masson N, Willam C, Maxwell PH, Pugh CW and Ratcliffe PJ . (2001). EMBO J., 20, 5197–5206.
Masuyama H and MacDonald PN . (1998). J. Cell Biochem., 71, 429–440.
Miao GG and Curran T . (1994). Mol. Cell. Biol., 14, 4295–4310.
Navon A and Goldberg AL . (2001). Mol. Cell, 8, 1339–1349.
Okazaki K and Sagata N . (1995). EMBO J., 14, 5048–5059.
Papavassiliou AG, Treier M, Chavrier C and Bohmann D . (1992). Science, 258, 1941–1944.
Perkins G, Drury LS and Diffley JF . (2001). EMBO J., 20, 4836–4845.
Pickart CM . (2001). Annu. Rev. Biochem., 70, 503–533.
Piechaczyk M and Blanchard JM . (1994). Crit. Rev. Oncol. Hematol., 17, 93–131.
Ransone LJ, Visvader J, Wamsley P and Verma IM . (1990). Proc. Natl. Acad. Sci. USA, 87, 3806–3810
Rech J and Fort P . (1989). Nucleic Acids Res., 17, 2874.
Rechsteiner M and Rogers SW . (1996). Trends Biochem. Sci., 21, 267–271.
Rogers S, Wells R and Rechsteiner M . (1986). Science, 234, 364–368.
Roux P, Blanchard JM, Fernandez A, Lamb N, Jeanteur P and Piechaczyk M . (1990). Cell, 63, 341–351
Rubio N, Rojo P and Torres C . (1996). J. Neurovirol., 2, 167–174.
Salvat C, Jariel-Encontre I, Acquaviva C, Omura S and Piechaczyk M . (1998). Oncogene, 17, 327–337.
Sambrook J, Fritsch EF and Maniatis T . (1989). Molecular cloning Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY.
Schreiber M, Baumann B, Cotten M, Angel P and Wagner EF . (1995). EMBO J., 14, 5338–5349.
Schuermann M, Neuberg M, Hunter JB, Jenuwein T, Ryseck RP, Bravo R and Muller R . (1989). Cell, 56, 507–516.
Shaulian E and Karin M . (2001). Oncogene, 20, 2390–2400.
Stancovski I, Gonen H, Orian A, Schwartz AL and Ciechanover A . (1995). Mol. Cell. Biol., 15, 7106–7116
Su K, Yang X, Roos MD, Paterson AJ and Kudlow JE . (2000). Biochem. J., 348 (Part 2), 281–289.
Tajima S and Aida Y . (2002). J. Virol., 76, 2557–2562.
Tansey WP . (2001). Genes Dev., 15, 1045–1050.
Thomas D and Tyers M . (2000). Curr. Biol., 10, R341–R343.
Tsurumi C, Ishida N, Tamura T, Kakizuka A, Nishida E, Okumura E, Kishimoto T, Inagaki M, Okazaki K, Sagata N et al. (1995). Mol. Cell. Biol., 15, 5682–5687.
van Dam H and Castellazzi M . (2001). Oncogene, 20, 2453–2464.
Wang W, Chevray PM and Nathans D . (1996). Proc. Natl. Acad. Sci. USA, 93, 8236–8240.
Wang ZQ, Ovitt C, Grigoriadis AE, Mohle-Steinlein U, Ruther U and Wagner EF . (1992). Nature 360, 741–745
Zhang J, Zhang D, McQuade JS, Behbehani M, Tsien JZ and Xu M . (2002). Nat. Genet., 30, 416–420.
Acknowledgements
This work was supported by grants from the CNRS, the Association de Recherche contre le Cancer and the Quality of Life Program of the European Community (contract QLG1-CT-2001-02026). PF is a recipient of the Ligue contre le Cancer. We are grafetul to Dr O Coux for stimulating discussions and to Dr P Travo for helpful advices concerning fluorescence miscroscope analysis.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Ferrara, P., Andermarcher, E., Bossis, G. et al. The structural determinants responsible for c-Fos protein proteasomal degradation differ according to the conditions of expression. Oncogene 22, 1461–1474 (2003). https://doi.org/10.1038/sj.onc.1206266
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.onc.1206266
Keywords
This article is cited by
-
Thymidylate synthase O-GlcNAcylation: a molecular mechanism of 5-FU sensitization in colorectal cancer
Oncogene (2022)
-
AP-1cFos/JunB/miR-200a regulate the pro-regenerative glial cell response during axolotl spinal cord regeneration
Communications Biology (2019)
-
The Ying and Yang of Adenosine A1 and A2A Receptors on ERK1/2 Activation in a Rat Model of Global Cerebral Ischemia Reperfusion Injury
Molecular Neurobiology (2018)
-
Recurrent rearrangements of FOS and FOSB define osteoblastoma
Nature Communications (2018)
-
c-Fos Protects Neurons Through a Noncanonical Mechanism Involving HDAC3 Interaction: Identification of a 21-Amino Acid Fragment with Neuroprotective Activity
Molecular Neurobiology (2016)