Abstract
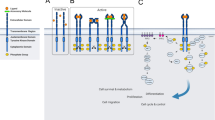
Tyrosine kinase oncogenes are formed as a result of mutations that induce constitutive kinase activity. Many of these tyrosine kinase oncogenes that are derived from genes, such as c-Abl, c-Fes, Flt3, c-Fms, c-Kit and PDGFRβ, that are normally involved in the regulation of hematopoiesis or hematopoietic cell function. Despite differences in structure, normal function, and subcellular location, many of the tyrosine kinase oncogenes signal through the same pathways, and typically enhance proliferation and prolong viability. They represent excellent potential drug targets, and it is likely that additional mutations will be identified in other kinases, their immediate downstream targets, or in proteins regulating their function.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Abe A, Emi N, Tanimoto M, Terasaki H, Marunouchi T, Saito H . 1997 Blood 90: 4271–4277
Abu-Duhier FM, Goodeve AC, Wilson GA, Care RS, Peake IR, Reilly JT . 2001 Br. J. Haematol. 113: 983–988
Afar DE, Han L, McLaughlin J, Wong S, Dhaka A, Parmar K, Rosenberg N, Witte ON, Colicelli J . 1997 Immunity 6: 773–782
Agami R, Blandino G, Oren M, Shaul Y . 1999 Nature 399: 809–813
Alai M, Mui AL, Cutler RL, Bustelo XR, Barbacid M, Krystal G . 1992 J. Biol. Chem. 267: 18021–18025
Alimandi M, Heidaran MA, Gutkind JS, Zhang J, Ellmore N, Valius M, Kazlauskas A, Pierce JH, Li W . 1997 Oncogene 15: 585–593
Amarante-Mendes GP, McGahon AJ, Nishioka WK, Afar DE, Witte ON, Green DR . 1998a Oncogene 16: 1383–1390
Amarante-Mendes GP, Naekyung Kim C, Liu L, Huang Y, Perkins CL, Green DR, Bhalla K . 1998b Blood 91: 1700–1705
Anderson SM, Jorgensen B . 1995 J. Immunol. 155: 1660–1670
Andreasson P, Johansson B, Carlsson M, Jarlsfeldt I, Fioretos T, Mitelman F, Hoglund M . 1997 Genes Chrom. Cancer 20: 299–304
Arceci RJ, Shanahan F, Stanley ER, Pollard JW . 1989 Proc. Natl. Acad. Sci. USA 86: 8818–8822
Armstrong SA, Staunton JE, Silverman LB, Pieters R, den Boer ML, Minden MD, Sallan SE, Lander ES, Golub TR, Korsmeyer SJ . 2002 Nat. Genet. 30: 41–47
Arvidsson AK, Rupp E, Nanberg E, Downward J, Ronnstrand L, Wennstrom S, Schlessinger J, Heldin CH, Claesson-Welsh L . 1994 Mol. Cell. Biol. 14: 6715–6726
Ashmun RA, Look AT, Roberts WM, Roussel MF, Seremetis S, Ohtsuka M, Sherr CJ . 1989 Blood 73: 827–837
Aziz N, Cherwinski H, McMahon M . 1999 Mol. Cell. Biol. 19: 1101–1115
Bai RY, Dieter P, Peschel C, Morris SW, Duyster J . 1998a Mol. Cell. Biol. 18: 6951–6961
Bai RY, Jahn T, Schrem S, Munzert G, Weidner KM, Wang JY, Duyster J . 1998b Oncogene 17: 941–948
Bai RY, Ouyang T, Miething C, Morris SW, Peschel C, Duyster J . 2000 Blood 96: 4319–4327
Barone MV, Courtneidge SA . 1995 Nature 378: 509–512
Baskaran R, Dahmus ME, Wang JY . 1993 Proc. Natl. Acad. Sci. USA 90: 11167–11171
Baskaran R, Wood LD, Whitaker LL, Canman CE, Morgan SE, Xu Y, Barlow C, Baltimore D, Wynshaw-Boris A, Kastan MB, Wang JY . 1997 Nature 387: 516–519
Baxter RM, Secrist JP, Vaillancourt RR, Kazlauskas A . 1998 J. Biol. Chem. 273: 17050–17055
Beghini A, Peterlongo P, Ripamonti CB, Larizza L, Cairoli R, Morra E, Mecucci C . 2000 Blood 95: 726–727
Benharroch D, Meguerian-Bedoyan Z, Lamant L, Amin C, Brugieres L, Terrier-Lacombe MJ, Haralambieva E, Pulford K, Pileri S, Morris SW, Mason DY, Delsol G . 1998 Blood 91: 2076–2084
Bergsten E, Uutela M, Li X, Pietras K, Ostman A, Heldin CH, Alitalo K, Eriksson U . 2001 Nat. Cell. Biol. 3: 512–516
Birchenall-Roberts MC, Ruscetti FW, Kasper JJ, Bertolette III DC, Yoo YD, Bang OS, Roberts MS, Turley JM, Ferris DK, Kim SJ . 1995 Mol. Cell. Biol. 15: 6088–6099
Birchenall-Roberts MC, Yoo YD, Bertolette DR, Lee KH, Turley JM, Bang OS, Ruscetti FW, Kim SJ . 1997 J. Biol. Chem. 272: 8905–8911
Birg F, Courcoul M, Rosnet O, Bardin F, Pebusque MJ, Marchetto S, Tabilio A, Mannoni P, Birnbaum D . 1992 Blood 80: 2584–2593
Bischof D, Pulford K, Mason DY, Morris SW . 1997 Mol. Cell. Biol. 17: 2312–2325
Bitter MA, Franklin WA, Larson RA, McKeithan TW, Rubin CM, Le Beau MM, Stephens JK, Vardiman JW . 1990 Am. J. Surg. Pathol. 14: 305–316
Blume-Jensen P, Hunter T . 2001 Nature 411: 355–365
Borer RA, Lehner CF, Eppenberger HM, Nigg EA . 1989 Cell 56: 379–390
Boultwood J, Rack K, Kelly S, Madden J, Sakaguchi AY, Wang LM, Oscier DG, Buckle VJ, Wainscoat JS . 1991 Proc. Natl. Acad. Sci. USA 88: 6176–6180
Bourette RP, De Sepulveda P, Arnaud S, Dubreuil P, Rottapel R, Mouchiroud G . 2001 J. Biol. Chem. 276: 22133–22139
Bourette RP, Myles GM, Carlberg K, Chen AR, Rohrschneider LR . 1995 Cell Growth Differ. 6: 631–645
Bourette RP, Rohrschneider LR . 2000 Growth Factors 17: 155–166
Brasel K, Escobar S, Anderberg R, de Vries P, Gruss HJ, Lyman SD . 1995 Leukemia 9: 1212–1218
Brasel K, McKenna HJ, Morrisey PJ, Charrier K, Morris AE, Lee CC, Williams DE, Lyman SD . 1996 Blood 88: 2004–2012
Bridge JA, Kanamori M, Ma Z, Pickering D, Hill DA, Lydiatt W, Lui MY, Colleoni GW, Antonescu CR, Ladanyi M, Morris SW . 2001 Am. J. Pathol. 159: 411–415
Brizzi MF, Aronica MG, Rosso A, Bagnara GP, Yarden Y, Pegoraro L . 1996a J. Biol. Chem. 271: 3562–3567
Brizzi MF, Dentelli P, Lanfrancone L, Rosso A, Pelicci PG, Pegoraro L . 1996b Oncogene 13: 2067–2076
Brizzi MF, Dentelli P, Rosso A, Yarden Y, Pegoraro L . 1999 J. Biol. Chem. 274: 16965–16972
Brosnan CF, Shafit-Zagardo B, Aquino DA, Berman JW . 1993 Adv. Neurol. 59: 349–361
Carlberg K, Rohrschneider LR . 1997 J. Biol. Chem. 272: 15943–15950
Carlesso N, Frank DA, Griffin JD . 1996 J. Exper. Med. 183: 811–820
Carow CE, Levenstein M, Kaufmann SH, Chen J, Amin S, Rockwell P, Witte L, Borowitz MJ, Civin CI, Small D . 1996 Blood 87: 1089–1096
Carpino N, Wisniewski D, Strife A, Marshak D, Kobayashi R, Stillman B, Clarkson B . 1997 Cell 88: 197–204
Caruana G, Cambareri AC, Ashman LK . 1999 Oncogene 18: 5573–5581
Cazzaniga G, Tosi S, Aloisi A, Giudici G, Daniotti M, Pioltelli P, Kearney L, Biondi A . 1999 Blood 94: 4370–4373
Chabot B, Stephenson DA, Chapman VM, Besmer P, Bernstein A . 1988 Nature 335: 88–89
Chen G, Yuan SS, Liu W, Xu Y, Trujillo K, Song B, Cong F, Goff SP, Wu Y, Arlinghaus R, Baltimore D, Gasser PJ, Park MS, Sung P, Lee EY . 1999 J. Biol. Chem. 274: 12748–12752
Chen K, Braun S, Lyman S, Fan Y, Traycoff CM, Wiebke EA, Gaddy J, Sledge G, Broxmeyer HE, Cornetta K . 1997 Cancer Res. 57: 3511–3516
Chen M, She H, Kim A, Woodley DT, Li W . 2000 Mol. Cell Biol. 20: 7867–7880
Cheng H, Rogers JA, Dunham NA, Smithgall TE . 1999 Mol. Cell. Biol. 19: 8335–8343
Cheng HY, Schiavone AP, Smithgall TE . 2001 Mol. Cell. Biol. 21: 6170–6180
Chian R, Young S, Danilkovitch-Miagkova A, Ronnstrand L, Leonard E, Ferrao P, Ashman L, Linnekin D . 2001 Blood 98: 1365–1373
Coffin CM, Dehner LP, Meis-Kindblom JM . 1998 Semin. Diagn. Pathol. 15: 102–110
Coffin CM, Patel A, Perkins S, Elenitoba-Johnson KS, Perlman E, Griffin CA . 2001 Mod. Pathol. 14: 569–576
Colleoni GW, Bridge JA, Garicochea B, Liu J, Filippa DA, Ladanyi M . 2000 Am. J. Pathol. 156: 781–789
Cong F, Yuan B, Goff SP . 1999 Mol. Cell. Biol. 19: 8314–8325
Cook JR, Dehner LP, Collins MH, Ma Z, Morris SW, Coffin CM, Hill DA . 2001 Am. J. Surg. Pathol. 25: 1364–1371
Coutinho S, Jahn T, Lewitzky M, Feller S, Hutzler P, Peschel C, Duyster J . 2000 Blood 96: 618–624
Csar XF, Wilson NJ, McMahon KA, Marks DC, Beecroft TL, Ward AC, Whitty GA, Kanangasundarum V, Hamilton JA . 2001 J. Biol. Chem. 276: 26211–26217
Dai XM, Ryan GR, Hapel AJ, Dominguez MG, Russell RG, Kapp S, Sylvestre V, Stanley ER . 2002 Blood 99: 111–120
Dai Z, Kerzic P, Schroeder WG, McNiece IK . 2001 J. Biol. Chem. 276: 28954–28960
Dai Z, Pendergast AM . 1995 Genes Dev. 9: 2569–2582
Danhauser-Riedl S, Warmuth M, Druker BJ, Emmerich B, Hallek M . 1996 Cancer Res. 56: 3589–3596
Daynes RA, Dowell T, Araneo BA . 1991 J. Exp. Med. 174: 1323–1333
de Jong R, ten Hoeve J, Heisterkamp N, Groffen J . 1995 J. Biol. Chem. 270: 21468–21471
de Jong R, van Wijk A, Haataja L, Heisterkamp N, Groffen J . 1997 J. Biol. Chem. 272: 32649–32655
de Parseval N, Fichelson S, Mayeux P, Gisselbrecht S, Sola B . 1993 Cytokine 5: 8–15
De Sepulveda P, Okkenhaug K, Rose JL, Hawley RG, Dubreuil P, Rottapel R . 1999 EMBO J. 18: 904–915
Delwiche F, Raines E, Powell J, Ross R, Adamson J . 1985 J. Clin. Invest. 76: 137–142
Dey A, She H, Kim L, Boruch A, Guris DL, Carlberg K, Sebti SM, Woodley DT, Imamoto A, Li W . 2000 Mol. Biol. Cell. 11: 3835–3848
Di Cristofano A, Carpino N, Dunant N, Friedland G, Kobayashi R, Strife A, Wisniewski D, Clarkson B, Pandolfi PP, Resh MD . 1998 J. Biol. Chem. 273: 4827–4830
Dickens M, Rogers JS, Cavanagh J, Raitano A, Xia Z, Halpern JR, Greenberg ME, Sawyers CL, Davis RJ . 1997 Science 277: 693–696
Dosil M, Wang S, Lemischka IR . 1993 Mol. Cell. Biol. 13: 6572–6585
Dubrez L, Eymin B, Sordet O, Droin N, Turhan AG, Solary E . 1998 Blood 91: 2415–2422
Duyster J, Baskaran R, Wang JY . 1995 Proc. Natl. Acad. Sci. USA 92: 1555–1559
Egan SE, Giddings BW, Brooks MW, Buday L, Sizeland AM, Weinberg RA . 1993 Nature 363: 45–51
Ernst TJ, Slattery KE, Griffin JD . 1994 J. Biol. Chem. 269: 5764–5769
Escalante M, Courtney J, Chin WG, Teng KK, Kim JI, Fajardo JE, Mayer BJ, Hempstead BL, Birge RB . 2000 J. Biol. Chem. 275: 24787–24797
Falini B, Bigerna B, Fizzotti M, Pulford K, Pileri SA, Delsol G, Carbone A, Paulli M, Magrini U, Menestrina F, Giardini R, Pilotti S, Mezzelani A, Ugolini B, Billi M, Pucciarini A, Pacini R, Pelicci PG, Flenghi L . 1998 Am. J. Pathol. 153: 875–886
Falini B, Pulford K, Pucciarini A, Carbone A, De Wolf-Peeters C, Cordell J, Fizzotti M, Santucci A, Pelicci PG, Pileri S, Campo E, Ott G, Delsol G, Mason DY . 1999 Blood 94: 3509–3515
Feldman RA, Wang E, Hanafusa H . 1983 J. Virol. 45: 782–791
Feller SM, Knudsen B, Hanafusa H . 1995 Oncogene 10: 1465–1473
Feng GS, Ouyang YB, Hu DP, Shi ZQ, Gentz R, Ni J . 1996 J. Biol. Chem. 271: 12129–12132
Fenski R, Flesch K, Serve S, Mizuki M, Oelmann E, Kratz-Albers K, Kienast J, Leo R, Schwartz S, Berdel WE, Serve H . 2000 Br. J. Haematol. 108: 322–330
Ferrari S, Manfredini R, Tagliafico E, Grande A, Barbieri D, Balestri R, Pizzanelli M, Zucchini P, Citro G, Zupi G, Franceschi C, Torelli U . 1994 Leukemia 8: Suppl 1 S91–S94
Fleischman RA, Saltman DL, Stastny V, Zneimer S . 1991 Proc. Natl. Acad. Sci. USA 88: 10885–10889
Foss B, Ulvestad E, Bruserud O . 2001 Eur. J. Haematol. 66: 365–376
Fowles LF, Martin ML, Nelsen L, Stacey KJ, Redd D, Clark YM, Nagamine Y, McMahon M, Hume DA, Ostrowski MC . 1998 Mol. Cell. Biol. 18: 5148–5156
Fujimoto J, Shiota M, Iwahara T, Seki N, Satoh H, Mori S, Yamamoto T . 1996 Proc. Natl. Acad. Sci. USA 93: 4181–4186
Gari M, Goodeve A, Wilson G, Winship P, Langabeer S, Linch D, Vandenberghe E, Peake I, Reilly J . 1999 Br. J. Haematol. 105: 894–900
Garrington TP, Ishizuka T, Papst PJ, Chayama K, Webb S, Yujiri T, Sun W, Sather S, Russell DM, Gibson SB, Keller G, Gelfand EW, Johnson GL . 2000 EMBO J. 19: 5387–5395
Gersuk GM, Westermark B, Mohabeer AJ, Challita PM, Pattamakom S, Pattengale PK . 1991 Scand. J. Immunol. 33: 521–532
Gesbert F, Griffin JD . 2000 Blood 96: 2269–2276
Gesbert F, Sellers WR, Signoretti S, Loda M, Griffin JD . 2000 J. Biol. Chem. 275: 39223–39230
Giebel LB, Spritz RA . 1991 Proc. Natl. Acad. Sci. USA 88: 8696–8699
Gisselbrecht S, Fichelson S, Sola B, Bordereaux D, Hampe A, Andre C, Galibert F, Tambourin P . 1987 Nature 329: 259–261
Golub TG, Barker G, Lovett M, Gilliland DG . 1994 Cell 77: 307–316
Golub TR, Goga A, Barker GF, Afar DE, McLaughlin J, Bohlander SK, Rowley JD, Witte ON, Gilliland DG . 1996 Mol. Cell. Biol. 16: 4107–4116
Gommerman JL, Berger SA . 1998 Blood 91: 1891–1900
Gommerman JL, Sittaro D, Klebasz NZ, Williams DA, Berger SA . 2000 Blood 96: 3734–3742
Gong JG, Costanzo A, Yang HQ, Melino G, Kaelin Jr WG, Levrero M, Wang JY . 1999 Nature 399: 806–809
Gotoh A, Miyazawa K, Ohyashiki K, Toyama K . 1994 Leukemia 8: 115–120
Greenland C, Touriol C, Chevillard G, Morris SW, Bai R, Duyster J, Delsol G, Allouche M . 2001 Oncogene 20: 7386–7397
Greer P, Haigh J, Mbamalu G, Khoo W, Bernstein A, Pawson T . 1994 Mol. Cell. Biol. 14: 6755–6763
Grey A, Chen Y, Paliwal I, Carlberg K, Insogna K . 2000 Endocrinology 141: 2129–2138
Griffin CA, Hawkins AL, Dvorak C, Henkle C, Ellingham T, Perlman EJ . 1999 Cancer Res. 59: 2776–2780
Groffen J, Heisterkamp N, Shibuya M, Hanafusa H, Stephenson JR . 1983 Virology 125: 480–486
Gross AW, Zhang X, Ren R . 1999 Mol. Cell. Biol. 19: 6918–6928
Hackenmiller R, Kim J, Feldman RA, Simon MC . 2000 Immunity 13: 397–407
Haigh J, McVeigh J, Greer P . 1996 Cell Growth Differ. 7: 931–944
Hanazono Y, Chiba S, Sasaki K, Mano H, Miyajima A, Arai K, Yazaki Y, Hirai H . 1993a EMBO J. 12: 1641–1646
Hanazono Y, Chiba S, Sasaki K, Mano H, Yazaki Y, Hirai H . 1993b Blood 81: 3193–3196
Hannum C, Culpepper J, Campbell D, McClanahan T, Zurawski S, Bazan JF, Kastelein R, Hudak S, Wagner J, Mattson J, Luh J, Duda G, Martina N, Peterson D, Menon S, Shanafelt A, Muench M, Kelner G, Namikawa R, Rennick D, Roncarolo M-G, Zlotnik A, Rosnet O, Dubreuil P, Birnbaum D, Lee F . 1994 Nature 368: 643–648
Hao SX, Ren R . 2000 Mol. Cell. Biol. 20: 1149–1161
Hardin JD, Boast S, Schwartzberg PL, Lee G, Alt FW, Stall AM, Goff SP . 1996 Cell Immunol. 172: 100–107
Hatch WC, Ganju RK, Hiregowdara D, Avraham S, Groopman JE . 1998 Blood 91: 3967–3973
Hernandez L, Pinyol M, Hernandez S, Bea S, Pulford K, Rosenwald A, Lamant L, Falini B, Ott G, Mason DY, Delsol G, Campo E . 1999 Blood 94: 3265–3268
Hirayama F, Lyman SD, Clark SC, Ogawa M . 1995 Blood 85: 1762–1768
Hjermstad SJ, Peters KL, Briggs SD, Glazer RI, Smithgall TE . 1993 Oncogene 8: 2283–2292
Honda H, Fujii T, Takatoku M, Mano H, Witte ON, Yazaki Y, Hirai H . 1995 Blood 85: 2853–2861
Honda H, Oda H, Suzuki T, Takahashi T, Witte ON, Ozawa K, Ishikawa T, Yazaki Y, Hirai H . 1998 Blood 91: 2067–2075
Hongyo T, Li T, Syaifudin M, Baskar R, Ikeda H, Kanakura Y, Aozasa K, Nomura T . 2000 Cancer Res. 60: 2345–2347
Huang E, Nocka K, Beier DR, Chu TY, Buck J, Lahm HW, Wellner D, Leder P, Besmer P . 1990 Cell 63: 225–233
Huber M, Helgason CD, Scheid MP, Duronio V, Humphries RK, Krystal G . 1998 EMBO J. 17: 7311–7319
Hudak S, Hunte B, Culpepper J, Menon S, Hannum C, Thompson-Snipes L, Rennick D . 1995 Blood 85: 2747–2755
Iijima Y, Ito T, Oikawa T, Eguchi M, Eguchi-Ishimae M, Kamada N, Kishi N, Asano S, Sakaki Y, Sato Y . 2000 Blood 95: 2126–2131
Ilaria Jr RL, Van Etten RA . 1996 J. Biol. Chem. 271: 31704–31710
Irusta PM, DiMaio D . 1998 EMBO J. 17: 6912–6923
Ito Y, Pandey P, Sathyanarayana P, Ling P, Rana A, Weichselbaum R, Tan TH, Kufe D, Kharbanda S . 2001 J. Biol. Chem. 276: 18130–18138
Iwahara T, Fujimoto J, Wen D, Cupples R, Bucay N, Arakawa T, Mori S, Ratzkin B, Yamamoto T . 1997 Oncogene 14: 439–449
Izuhara K, Feldman RA, Greer P, Harada N . 1994 J. Biol. Chem. 269: 18623–18629
Jacobsen SE, Okkenhaug C, Myklebust J, Veiby OP, Lyman SD . 1995 J. Exp. Med. 181: 1357–1363
Jain SK, Langdon WY, Varticovski L . 1997 Oncogene 14: 2217–2228
Jiang H, Foltenyi K, Kashiwada M, Donahue L, Vuong B, Hehn B, Rothman P . 2001 J. Immunol. 166: 2627–2634
Jonuleit T, van der Kuip H, Miething C, Michels H, Hallek M, Duyster J, Aulitzky WE . 2000 Blood 96: 1933–1939
Jucker M, McKenna K, da Silva AJ, Rudd CE, Feldman RA . 1997 J. Biol. Chem. 272: 2104–2109
Jucker M, Roebroek AJ, Mautner J, Koch K, Eick D, Diehl V, Van de Ven WJ, Tesch H . 1992 Oncogene 7: 943–952
Kadlec L, Pendergast AM . 1997 Proc. Natl. Acad. Sci. USA 94: 12390–12395
Kain KH, Klemke RL . 2001 J. Biol. Chem. 276: 16185–16192
Kaminski WE, Lindahl P, Lin NL, Broudy VC, Crosby JR, Hellstrom M, Swolin B, Bowen-Pope DF, Martin PJ, Ross R, Betsholtz C, Raines EW . 2001 Blood 97: 1990–1998
Karlsson T, Songyang Z, Landgren E, Lavergne C, Di Fiore PP, Anafi M, Pawson T, Cantley LC, Claesson-Welsh L, Welsh M . 1995 Oncogene 10: 1475–1483
Kashishian A, Kazlauskas A, Cooper JA . 1992 EMBO J. 11: 1373–1382
Kelly LM, Liu Q, Kutok JL, Williams IR, Boulton CL, Gilliland DG . 2002 Blood 99: 310–318
Kharbanda S, Pandey P, Yamauchi T, Kumar S, Kaneki M, Kumar V, Bharti A, Yuan ZM, Ghanem L, Rana A, Weichselbaum R, Johnson G, Kufe D . 2000 Mol. Cell. Biol. 20: 4979–4989
Kharbanda S, Ren R, Pandey P, Shafman TD, Feller SM, Weichselbaum RR, Kufe DW . 1995 Nature 376: 785–788
Kin Y, Li G, Shibuya M, Maru Y . 2001 J. Biol. Chem. 13: 13–
Kiyoi H, Naoe T, Yokota S, Nakao M, Minami S, Kuriyama K, Takeshita A, Saito K, Hasegawa S, Shimodaira S, Tamura J, Shimazaki C, Matsue K, Kobayashi H, Arima N, Suzuki R, Morishita H, Saito H, Ueda R, Ohno R . 1997 Leukemia 11: 1447–1452
Koleske AJ, Gifford AM, Scott ML, Nee M, Bronson RT, Miczek KA, Baltimore D . 1998 Neuron 21: 1259–1272
Kozlowski M, Larose L, Lee F, Le DM, Rottapel R, Siminovitch KA . 1998 Mol. Cell. Biol. 18: 2089–2099
Kruh GD, Perego R, Miki T, Aaronson SA . 1990 Proc. Natl. Acad. Sci. USA 87: 5802–5806
Kubota Y, Angelotti T, Niederfellner G, Herbst R, Ullrich A . 1998 Cell Growth Differ. 9: 247–256
Kuefer MU, Look AT, Pulford K, Behm FG, Pattengale PK, Mason DY, Morris SW . 1997 Blood 90: 2901–2910
Kulkarni S, Heath C, Parker S, Chase A, Iqbal S, Pocock CF, Kaeda J, Cwynarski K, Goldman JM, Cross NC . 2000 Cancer Res. 60: 3592–3598
Lagasse E, Weissman IL . 1997 Cell 89: 1021–1031
Lamant L, Dastugue N, Pulford K, Delsol G, Mariame B . 1999 Blood 93: 3088–3095
LaRochelle WJ, Jeffers M, McDonald WF, Chillakuru RA, Giese NA, Lokker NA, Sullivan C, Boldog FL, Yang M, Vernet C, Burgess CE, Fernandes E, Deegler LL, Rittman B, Shimkets J, Shimkets RA, Rothberg JM, Lichenstein HS . 2001 Nat. Cell. Biol. 3: 517–521
Lavagna-Sevenier C, Marchetto S, Birnbaum D, Rosnet O . 1998a J. Biol. Chem. 273: 14962–14967
Lavagna-Sevenier C, Marchetto S, Birnbaum D, Rosnet O . 1998b Leukemia 12: 301–310
Law SF, Estojak J, Wang B, Mysliwiec T, Kruh G, Golemis EA . 1996 Mol. Cell. Biol. 16: 3327–3337
Lawrence B, Perez-Atayde A, Hibbard MK, Rubin BP, Dal Cin P, Pinkus JL, Pinkus GS, Xiao S, Yi ES, Fletcher CD, Fletcher JA . 2000 Am. J. Pathol. 157: 377–384
Lee AW, States DJ . 2000 Mol. Cell. Biol. 20: 6779–6798
Lee PS, Wang Y, Dominguez MG, Yeung YG, Murphy MA, Bowtell DD, Stanley ER . 1999 EMBO J. 18: 3616–3628
Lewis JM, Schwartz MA . 1998 J. Biol. Chem. 273: 14225–14230
Li J, Smithgall TE . 1996 J. Biol. Chem. 271: 32930–32936
Li J, Smithgall TE . 1998 J. Biol. Chem. 273: 13828–13834
Li S, Couvillon AD, Brasher BB, Van Etten RA . 2001 EMBO J. 20: 6793–6804
Li S, Ilaria RLJ, Million RP, Daley GQ, Van Etten RA . 1999 J. Exp. Med. 189: 1399–1412
Li X, Ponten A, Aase K, Karlsson L, Abramsson A, Uutela M, Backstrom G, Hellstrom M, Bostrom H, Li H, Soriano P, Betsholtz C, Heldin CH, Alitalo K, Ostman A, Eriksson U . 2000 Nat. Cell. Biol. 2: 302–309
Lindahl P, Hellstrom M, Kalen M, Karlsson L, Pekny M, Pekna M, Soriano P, Betsholtz C . 1998 Development 125: 3313–3322
Linnekin D, DeBerry CS, Mou S . 1997 J. Biol. Chem. 272: 27450–27455
Linnekin D, Mou SM, Greer P, Longo DL, Ferris DK . 1995 J. Biol. Chem. 270: 4950–4954
Lioubin MN, Algate PA, Tsai S, Carlberg K, Aebersold A, Rohrschneider LR . 1996 Genes Dev. 10: 1084–1095
Lisovsky M, Estrov Z, Zhang X, Consoli U, Sanchez-Williams G, Snell V, Munker R, Goodacre A, Savchenko V, Andreeff M . 1996 Blood 88: 3987–3997
Liu SK, McGlade CJ . 1998 Oncogene 17: 3073–3082
Liu Y, Jenkins B, Shin JL, Rohrschneider LR . 2001 Mol. Cell. Biol. 21: 3047–3056
Longley Jr BJ, Metcalfe DD, Tharp M, Wang X, Tyrrell L, Lu SZ, Heitjan D, Ma Y . 1999 Proc. Natl. Acad. Sci. USA 96: 1609–1614
Longley BJ, Tyrrell L, Lu SZ, Ma YS, Langley K, Ding TG, Duffy T, Jacobs P, Tang LH, Modlin I . 1996 Nat. Genet. 12: 312–314
Lu L, Osmond DG . 2001 Exp. Hematol. 29: 596–601
Lu-Kuo JM, Fruman DA, Joyal DM, Cantley LC, Katz HR . 2000 J. Biol. Chem. 275: 6022–6029
Lyman SD, Jacobsen SE . 1998 Blood 91: 1101–1134
Lyman SD, James L, Vanden Bos T, de Vries P, Brasel K, Gliniak B, Hollingsworth LT, Picha KS, McKenna HJ, Splett RR, Fletcher FA, Maraskovsky E, Farrah T, Foxworthe D, Williams DE, Beckmann MP . 1993 Cell 75: 1157–1167
Lynch DH, Andreasen A, Maraskovsky E, Whitmore J, Miller RE, Schuh JC . 1997 Nat. Med. 3: 625–631
Ma Z, Cools J, Marynen P, Cui X, Siebert R, Gesk S, Schlegelberger B, Peeters B, De Wolf-Peeters C, Wlodarska I, Morris SW . 2000 Blood 95: 2144–2149
MacDonald I, Levy J, Pawson T . 1985 Mol. Cell. Biol. 5: 2543–2551
Mackarehtschian K, Hardin JD, Moore KA, Boast S, Goff SP, Lemischka IR . 1995 Immunity 3: 147–161
Magnusson MK, Meade KE, Brown KE, Arthur DC, Krueger LA, Barrett AJ, Dunbar CE . 2001 Blood 98: 2518–2525
Manfredini R, Balestri R, Tagliafico E, Trevisan F, Pizzanelli M, Grande A, Barbieri D, Zucchini P, Citro G, Franceschi C, Ferrari S . 1997 Blood 89: 135–145
Manfredini R, Grande A, Tagliafico E, Barbieri D, Zucchini P, Citro G, Zupi G, Franceschi C, Torelli U, Ferrari S . 1993 J. Exp. Med. 178: 381–389
Maraskovsky E, Brasel K, Teepe M, Roux ER, Lyman SD, Shortman K, McKenna HJ . 1996 J. Exp. Med. 184: 1953–1962
Maraskovsky E, Daro E, Roux E, Teepe M, Maliszewski CR, Hoek J, Caron D, Lebsack ME, McKenna HJ . 2000 Blood 96: 878–884
Marchetto S, Fournier E, Beslu N, Aurran-Schleinitz T, Dubreuil P, Borg JP, Birnbaum D, Rosnet O . 1999 Leukemia 13: 1374–1382
Maru Y, Peters KL, Afar DE, Shibuya M, Witte ON, Smithgall TE . 1995 Mol. Cell. Biol. 15: 835–842
Mason DY, Bastard C, Rimokh R, Dastugue N, Huret JL, Kristofferson U, Magaud JP, Nezelof C, Tilly H, Vannier JP et al . 1990 Br. J. Haematol. 74: 161–168
Mason DY, Pulford KA, Bischof D, Kuefer MU, Butler LH, Lamant L, Delsol G, Morris SW . 1998 Cancer Res. 58: 1057–1062
Matsuda T, Fukada T, Takahashi-Tezuka M, Okuyama Y, Fujitani Y, Hanazono Y, Hirai H, Hirano T . 1995 J. Biol. Chem. 270: 11037–11039
Matsuguchi T, Inhorn RC, Carlesso N, Xu G, Druker B, Griffin JD . 1995 EMBO J. 14: 257–265
Matsui T, Heidaran M, Miki T, Popescu N, La Rochelle W, Kraus M, Pierce J, Aaronson S . 1989 Science 243: 800–804
Matthews W, Jordan CT, Wiegand GW, Pardoll D, Lemischka IR . 1991 Cell 65: 1143–1152
McClanahan T, Culpepper J, Campbell D, Wagner J, Franz-Bacon K, Mattson J, Tsai S, Luh J, Guimaraes MJ, Mattei MG, Rosnet O, Birnbaum D, Hannum CH . 1996 Blood 88: 3371–3382
McGlynn H, Kapelko K, Baker A, Burnett A, Padua RA . 1997 Leuk. Res. 21: 919–923
McKenna HJ, Smith FO, Brasel K, Hirschstein D, Bernstein ID, Williams DE, Lyman SD . 1996 Exp. Hematol. 24: 378–385
McKenna HJ, Stocking KL, Miller RE, Brasel K, De Smedt T, Maraskovsky E, Maliszewski CR, Lynch DH, Smith J, Pulendran B, Roux ER, Teepe M, Lyman SE, Peschon JJ . 2000 Blood 95: 3489–3497
McMahon KA, Wilson NJ, Marks DC, Beecroft TL, Whitty GA, Hamilton JA, Csar XF . 2001 Biochem. J. 358: 431–436
McWhirter JR, Galasso DL, Wang JY . 1993 Mol. Cell. Biol. 13: 7587–7595
Meckling-Gill KA, Yee SP, Schrader JW, Pawson T . 1992 Biochim. Biophys. Acta 1137: 65–72
Meech SJ, McGavran L, Odom LF, Liang X, Meltesen L, Gump J, Wei Q, Carlsen S, Hunger SP . 2001 Blood 98: 1209–1216
Meierhoff G, Dehmel U, Gruss HJ, Rosnet O, Birnbaum D, Quentmeier H, Dirks W, Drexler HG . 1995 Leukemia 9: 1368–1372
Meshinchi S, Woods WG, Stirewalt DL, Sweetser DA, Buckley JD, Tjoa TK, Bernstein ID, Radich JP . 2001 Blood 97: 89–94
Michaelson MD, Bieri PL, Mehler MF, Xu H, Arezzo JC, Pollard JW, Kessler JA . 1996 Development 122: 2661–2672
Mori S, Ronnstrand L, Yokote K, Engstrom A, Courtneidge SA, Claesson-Welsh L, Heldin CH . 1993 EMBO J. 12: 2257–2264
Morris SW, Kirstein MN, Valentine MB, Dittmer KG, Shapiro DN, Saltman DL, Look AT . 1994 Science 263: 1281–1284
Morris SW, Naeve C, Mathew P, James PL, Kirstein MN, Cui X, Witte DP . 1997 Oncogene 14: 2175–2188
Morris SW, Xue L, Ma Z, Kinney MC . 2001 Br. J. Haematol. 113: 275–295
Nagata H, Worobec AS, Oh CK, Chowdhury BA, Tannenbaum S, Suzuki Y, Metcalfe DD . 1995 Proc. Natl. Acad. Sci. USA 92: 10560–10564
Nakao M, Yokota S, Iwai T, Kaneko H, Horiike S, Kashima K, Sonoda Y, Fujimoto T, Misawa S . 1996 Leukemia 10: 1911–1918
Nakata Y, Kimura A, Katoh O, Kawaishi K, Hyodo H, Abe K, Kuramoto A, Satow Y . 1995 Br. J. Haematol. 91: 661–663
Nelson KL, Rogers JA, Bowman TL, Jove R, Smithgall TE . 1998 J. Biol. Chem. 273: 7072–7077
Neshat MS, Raitano AB, Wang HG, Reed JC, Sawyers CL . 2000 Mol. Cell. Biol. 20: 1179–1186
Nicholls SE, Winter S, Mottram R, Miyan JA, Whetton AD . 1999 Exp. Hematol. 27: 663–672
Nieborowska-Skorska M, Slupianek A, Xue L, Zhang Q, Raghunath PN, Hoser G, Wasik MA, Morris SW, Skorski T . 2001 Cancer Res. 61: 6517–6523
Ning ZQ, Li J, Arceci RJ . 2001a Leuk. Lymphoma 41: 513–522
Ning ZQ, Li J, Arceci RJ . 2001b Blood 97: 3559–3567
Nishida K, Yoshida Y, Itoh M, Fukada T, Ohtani T, Shirogane T, Atsumi T, Takahashi-Tezuka M, Ishihara K, Hibi M, Hirano T . 1999 Blood 93: 1809–1816
Nishimura R, Li W, Kashishian A, Mondino A, Zhou M, Cooper J, Schlessinger J . 1993 Mol. Cell. Biol. 13: 6889–6896
Oda T, Heaney C, Hagopian JR, Okuda K, Griffin JD, Druker BJ . 1994 J. Biol. Chem. 269: 22925–22928
Pandey P, Raingeaud J, Kaneki M, Weichselbaum R, Davis RJ, Kufe D, Kharbanda S . 1996 J. Biol. Chem. 271: 23775–23779
Papadopoulos P, Ridge SA, Boucher CA, Stocking C, Wiedemann LM . 1995 Cancer Res. 55: 34–38
Parada Y, Banerji L, Glassford J, Lea NC, Collado M, Rivas C, Lewis JL, Gordon MY, Thomas NS, Lam EW . 2001 J. Biol. Chem. 276: 23572–23580
Park WY, Ahn JH, Feldman RA, Seo JS . 1998 Cancer Lett. 129: 29–37
Pear WS, Miller JP, Xu L, Pui JC, Soffer B, Quackenbush RC, Pendergast AM, Bronson R, Aster JC, Scott ML, Baltimore D . 1998 Blood 92: 3780–3792
Pendergast AM, Muller AJ, Havlik MH, Maru Y, Witte ON . 1991 Cell 66: 161–171
Pendergast AM, Quilliam LA, Cripe LD, Bassing CH, Dai Z, Li N, Batzer A, Rabun KM, Der CJ, Schlessinger J, Gishizky ML . 1993 Cell 75: 175–185
Perrotti D, Cesi V, Trotta R, Guerzoni C, Santilli G, Campbell K, Iervolino A, Condorelli F, Gambacorti-Passerini C, Caligiuri MA, Calabretta B . 2002 Nat. Genet. 30: 48–58
Piacibello W, Gammaitoni L, Bruno S, Gunetti M, Fagioli F, Cavalloni G, Aglietta M . 2000 J. Hematother. Stem. Cell. Res. 9: 945–956
Plattner R, Kadlec L, DeMali KA, Kazlauskas A, Pendergast AM . 1999 Genes Dev. 13: 2400–2411
Pollard JW, Hunt JS, Wiktor-Jedrzejczak W, Stanley ER . 1991 Dev. Biol. 148: 273–283
Price DJ, Rivnay B, Fu Y, Jiang S, Avraham S, Avraham H . 1997 J. Biol. Chem. 272: 5915–5920
Puil L, Liu J, Gish G, Mbamalu G, Bowtell D, Pelicci TG, Arlinghaus R, Pawson T . 1994 EMBO J. 13: 764–773
Pulendran B, Banchereau J, Burkeholder S, Kraus E, Guinet E, Chalouni C, Caron D, Maliszewski C, Davoust J, Fay J, Palucka K . 2000 J. Immunol. 165: 566–572
Pulford K, Falini B, Cordell J, Rosenwald A, Ott G, Muller-Hermelink HK, MacLennan KA, Lamant L, Carbone A, Campo E, Mason DY . 1999 Am. J. Pathol. 154: 1657–1663
Qi JH, Ito N, Claesson-Welsh L . 1999 J. Biol. Chem. 274: 14455–14463
Quelle FW, Sato N, Witthuhn BA, Inhorn RC, Eder M, Miyajima A, Griffin JD, Ihle JN . 1994 Mol. Cell. Biol. 14: 4335–4341
Raitano AB, Halpern JR, Hambuch TM, Sawyers CL . 1995 Proc. Natl. Acad. Sci. USA 92: 11746–11750
Rambaldi A, Wakamiya N, Vellenga E, Horiguchi J, Warren MK, Kufe D, Griffin JD . 1988 J. Clin. Invest. 81: 1030–1035
Rao P, Mufson RA . 1995 J. Biol. Chem. 270: 6886–6893
Reith AD, Ellis C, Lyman SD, Anderson DM, Williams DE, Bernstein A, Pawson T . 1991 EMBO J. 10: 2451–2459
Reuther JY, Reuther GW, Cortez D, Pendergast AM, Baldwin Jr AS . 1998 Genes Dev. 12: 968–981
Ridge SA, Worwood M, Oscier D, Jacobs A, Padua RA . 1990 Proc. Natl. Acad. Sci. USA 87: 1377–1380
Ritchie KA, Aprikyan AA, Bowen-Pope DF, Norby-Slycord CJ, Conyers S, Bartelmez S, Sitnicka EH, Hickstein DD . 1999 Leukemia 13: 1790–1803
Roebroek AJ, Schalken JA, Verbeek JS, Van den Ouweland AM, Onnekink C, Bloemers HP, Van de Ven WJ . 1985 EMBO J. 4: 2897–2903
Rogers JA, Cheng HY, Smithgall TE . 2000 Cell Growth Differ. 11: 581–592
Rogers JA, Read RD, Li J, Peters KL, Smithgall TE . 1996 J. Biol. Chem. 271: 17519–17525
Rombouts WJ, Blokland I, Lowenberg B, Ploemacher RE . 2000 Leukemia 14: 675–683
Ronnstrand L, Arvidsson AK, Kallin A, Rorsman C, Hellman U, Engstrom U, Wernstedt C, Heldin CH . 1999 Oncogene 18: 3696–3702
Rosnet O, Marchetto S, deLapeyriere O, Birnbaum D . 1991 Oncogene 6: 1641–1650
Rosnet O, Schiff C, Pebusque MJ, Marchetto S, Tonnelle C, Toiron Y, Birg F, Birnbaum D . 1993 Blood 82: 1110–1119
Ross TS, Bernard OA, Berger R, Gilliland DG . 1998 Blood 91: 4419–4426
Rottapel R, Turck CW, Casteran N, Liu X, Birnbaum D, Pawson T, Dubreuil P . 1994 Oncogene 9: 1755–1765
Roumiantsev S, de Aos IE, Varticovski L, Ilaria RL, Van Etten RA . 2001 Blood 97: 4–13
Roussel MF, Shurtleff SA, Downing JR, Sherr CJ . 1990 Proc. Natl. Acad. Sci. USA 87: 6738–6742
Sachsenmaier C, Sadowski HB, Cooper JA . 1999 Oncogene 18: 3583–3592
Saito Y, Haendeler J, Hojo Y, Yamamoto K, Berk BC . 2001 Mol. Cell. Biol. 21: 6387–6394
Salgia R, Pisick E, Sattler M, Li JL, Uemura N, Wong WK, Burky SA, Hirai H, Chen LB, Griffin JD . 1996a J. Biol. Chem. 271: 25198–25203
Salgia R, Sattler M, Pisick E, Li JL, Griffin JD . 1996b Exper. Hematol. 24: 310–331
Salgia R, Uemura N, Okuda K, Li JL, Pisick E, Sattler M, de Jong R, Druker B, Heisterkamp N, Chen LB, Griffin JD . 1995 J. Biol. Chem. 270: 29145–29150
Sanchez-Garcia I, Grutz G . 1995 Proc. Natl. Acad. Sci. USA 92: 5287–5291
Sattler M, Salgia R, Okuda K, Uemura N, Durstin MA, Pisick E, Xu G, Li JL, Prasad KV, Griffin JD . 1996 Oncogene 12: 839–846
Sattler M, Salgia R, Shrikhande G, Verma S, Choi JL, Rohrschneider LR, Griffin JD . 1997a Oncogene 15: 2379–2384
Sattler M, Salgia R, Shrikhande G, Verma S, Pisick E, Prasad KV, Griffin JD . 1997b J. Biol. Chem. 272: 10248–10253
Sattler M, Verma S, Pride YB, Salgia R, Rohrschneider LR, Griffin JD . 2001 J. Biol. Chem. 276: 2451–2458
Savvides SN, Boone T, Karplus AP . 2000 Nat. Struct. Biol. 7: 486–491
Sawyers CL, Callahan W, Witte ON . 1992 Cell 70: 901–910
Sawyers CL, McLaughlin J, Goga A, Havlik M, Witte O . 1994 Cell 77: 121–131
Sawyers CL, McLaughlin J, Witte ON . 1995 J. Exper. Med. 181: 307–313
Schwaller J, Anastasiadou E, Cain D, Kutok J, Wojiski S, Williams IR, LaStarza R, Crescenzi B, Sternberg DW, Andreasson P, Schiavo R, Siena S, Mecucci C, Gilliland DG . 2001 Blood 97: 3910–3918
Schwartzberg PL, Stall AM, Hardin JD, Bowdish KS, Humaran T, Boast S, Harbison ML, Robertson EJ, Goff SP . 1991 Cell 65: 1165–1175
Serve H, Yee NS, Stella G, Sepp-Lorenzino L, Tan JC, Besmer P . 1995 EMBO J. 14: 473–483
Sexl V, Piekorz R, Moriggl R, Rohrer J, Brown MP, Bunting KD, Rothammer K, Roussel MF, Ihle JN . 2000 Blood 96: 2277–2283
Shafman T, Khanna KK, Kedar P, Spring K, Kozlov S, Yen T, Hobson K, Gatei M, Zhang N, Watters D, Egerton M, Shiloh Y, Kharbanda S, Kufe D, Lavin MF . 1997 Nature 387: 520–523
Shi CS, Tuscano J, Kehrl JH . 2000 Blood 95: 776–782
Shi Y, Alin K, Goff SP . 1995 Genes Dev. 9: 2583–2597
Shiota M, Fujimoto J, Semba T, Satoh H, Yamamoto T, Mori S . 1994 Oncogene 9: 1567–1574
Siebert R, Gesk S, Harder L, Steinemann D, Grote W, Schlegelberger B, Tiemann M, Wlodarska I, Schemmel V . 1999 Blood 94: 3614–3617
Sillaber C, Gesbert F, Frank DA, Sattler M, Griffin JD . 2000 Blood 95: 2118–2125
Sionov RV, Coen S, Goldberg Z, Berger M, Bercovich B, Ben-Neriah Y, Ciechanover A, Haupt Y . 2001 Mol. Cell. Biol. 21: 5869–5878
Skorski T, Bellacosa A, Nieborowska-Skorska M, Majewski M, Martinez R, Choi JK, Trotta R, Wlodarski P, Perrotti D, Chan TO, Wasik MA, Tsichlis PN, Calabretta B . 1997 EMBO J. 16: 6151–6161
Skorski T, Kanakaraj P, Nieborowska-Skorska M, Ratajczak MZ, Wen SC, Zon G, Gewirtz AM, Perussia B, Calabretta B . 1995a Blood 86: 726–736
Skorski T, Nieborowska-Skorska M, Szczylik C, Kanakaraj P, Perrotti D, Zon G, Gewirtz A, Perussia B, Calabretta B . 1995b Cancer Res. 55: 2275–2278
Slupianek A, Nieborowska-Skorska M, Hoser G, Morrione A, Majewski M, Xue L, Morris SW, Wasik MA, Skorski T . 2001a Cancer Res. 61: 2194–2199
Slupianek A, Schmutte C, Tombline G, Nieborowska-Skorska M, Hoser G, Nowicki MO, Pierce AJ, Fishel R, Skorski T . 2001b Mol. Cell. 8: 795–806
Small D, Levenstein M, Kim E, Carow C, Amin S, Rockwell P, Witte L, Burrow C, Ratajczak MZ, Gewirtz AM, Givin CI . 1994 Proc. Natl. Acad. Sci. USA 91: 459–463
Smithgall TE, Johnston JB, Bustin M, Glazer RI . 1991 J. Natl. Cancer Inst. 83: 42–46
Smithgall TE, Yu G, Glazer RI . 1988 J. Biol. Chem. 263: 15050–15055
Sonoyama J, Matsumura I, Ezoe S, Satoh Y, Zhang X, Kataoka Y, Takai E, Mizuki M, Machii T, Wakao H, Kanakura Y . 2002 J. Biol. Chem. 4: 4–
Soriano P . 1994 Genes Dev. 8: 1888–1896
Stanley ER, Guilbert LJ, Tushinski RJ, Bartelmez SH . 1983 J. Cell. Biochem. 21: 151–159
Stein H, Foss HD, Durkop H, Marafioti T, Delsol G, Pulford K, Pileri S, Falini B . 2000 Blood 96: 3681–3695
Sternberg DW, Tomasson MH, Carroll M, Curley DP, Barker G, Caprio M, Wilbanks A, Kazlauskas A, Gilliland DG . 2001 Blood 98: 3390–3397
Sun X, Majumder P, Shioya H, Wu F, Kumar S, Weichselbaum R, Kharbanda S, Kufe D . 2000a J. Biol. Chem. 275: 17237–17240
Sun X, Wu F, Datta R, Kharbanda S, Kufe D . 2000b J. Biol. Chem. 275: 7470–7473
Sundberg C, Rubin K . 1996 J. Cell. Biol. 132: 741–752
Tallquist MD, Klinghoffer RA, Heuchel R, Mueting-Nelsen PF, Corrin PD, Heldin CH, Johnson RJ, Soriano P . 2000 Genes Dev. 14: 3179–3190
Tamura T, Mancini A, Joos H, Koch A, Hakim C, Dumanski J, Weidner KM, Niemann H . 1999 Oncogene 18: 6488–6495
Tauchi T, Boswell HS, Leibowitz D, Broxmeyer HE . 1994 J. Exper. Med. 179: 167–175
Tauchi T, Miyazawa K, Feng GS, Broxmeyer HE, Toyama K . 1997 J. Biol. Chem. 272: 1389–1394
Theis S, Roemer K . 1998 Oncogene 17: 557–564
Thommes K, Lennartson J, Carlberg M, Ronnstrand L . 1999 Biochem. J. 341: 211–216
Tian Q, Frierson Jr HF, Krystal GW, Moskaluk CA . 1999 Am. J. Pathol. 154: 1643–1647
Timokhina I, Kissel H, Stella G, Besmer P . 1998 EMBO J. 17: 6250–6262
Tobal K, Pagliuca A, Bhatt B, Bailey N, Layton DM, Mufti GJ . 1990 Leukemia 4: 486–489
Tomasson MH, Sternberg DW, Williams IR, Carroll M, Cain D, Aster JC, Ilaria Jr RL, Van Etten RA, Gilliland DG . 2000 J. Clin. Invest. 105: 423–432
Tomasson MH, Williams IR, Hasserjian R, Udomsakdi C, McGrath SM, Schwaller J, Druker B, Gilliland DG . 1999 Blood 93: 1707–1714
Tort F, Pinyol M, Pulford K, Roncador G, Hernandez L, Nayach I, Kluin-Nelemans HC, Kluin P, Touriol C, Delsol G, Mason D, Campo E . 2001 Lab. Invest. 81: 419–426
Touriol C, Greenland C, Lamant L, Pulford K, Bernard F, Rousset T, Mason DY, Delsol G . 2000 Blood 95: 3204–3207
Trinei M, Lanfrancone L, Campo E, Pulford K, Mason DY, Pelicci PG, Falini B . 2000 Cancer Res. 60: 793–798
Tybulewicz VL, Crawford CE, Jackson PK, Bronson RT, Mulligan RC . 1991 Cell 65: 1153–1163
Valgeirsdottir S, Paukku K, Silvennoinen O, Heldin CH, Claesson-Welsh L . 1998 Oncogene 16: 505–515
Valius M, Secrist JP, Kazlauskas A . 1995 Mol. Cell. Biol. 15: 3058–3071
van Dijk TB, van Den Akker E, Amelsvoort MP, Mano H, Lowenberg B, von Lindern M . 2000 Blood 96: 3406–3413
Van Etten RA, Jackson PK, Baltimore D, Sanders MC, Matsudaira PT, Janmey PA . 1994 J. Cell Biol. 124: 325–340
Vignais ML, Sadowski HB, Watling D, Rogers NC, Gilman M . 1996 Mol. Cell. Biol. 16: 1759–1769
Voncken JW, Kaartinen V, Pattengale PK, Germeraad WT, Groffen J, Heisterkamp N . 1995 Blood 86: 4603–4611
Vosseller K, Stella G, Yee NS, Besmer P . 1997 Mol. Biol. Cell 8: 909–922
Wang B, Golemis EA, Kruh GD . 1997 J. Biol. Chem. 272: 17542–17550
Wang B, Kruh GD . 1996 Oncogene 13: 193–197
Wang Y, Yeung YG, Langdon WY, Stanley ER . 1996 J. Biol. Chem. 271: 17–20
Weiss A., Schlessinger J . 1998 Cell 94: 277–280
Welch PJ, Wang JY . 1993 Cell 75: 779–790
Welch PJ, Wang JY . 1995 Mol. Cell. Biol. 15: 5542–5551
Wellmann A, Otsuki T, Vogelbruch M, Clark HM, Jaffe ES, Raffeld M . 1995 Blood 86: 2321–2328
Wen ST, Jackson PK, Van Etten RA . 1996 EMBO J. 15: 1583–1595
Whang YE, Tran C, Henderson C, Syljuasen RG, Rozengurt N, McBride WH, Sawyers CL . 2000 Proc. Natl. Acad. Sci. USA 97: 5486–5491
Whitman SP, Archer KJ, Feng L, Baldus C, Becknell B, Carlson BD, Carroll AJ, Mrozek K, Vardiman JW, George SL, Kolitz JE, Larson RA, Bloomfield CD, Caligiuri MA . 2001 Cancer Res. 61: 7233–7239
Wiktor-Jedrzejczak W, Bartocci A, Ferrante Jr AW, Ahmed-Ansari A, Sell KW, Pollard JW, Stanley ER . 1990 Proc. Natl. Acad. Sci. USA 87: 4828–4832
Wisniewski D, Strife A, Swendeman S, Erdjument-Bromage H, Geromanos S, Kavanaugh WM, Tempst P, Clarkson B . 1999 Blood 93: 2707–2720
Witthuhn BA, Quelle FW, Silvennoinen O, Yi T, Tang B, Miura O, Ihle JN . 1993 Cell 74: 227–236
Wlodarska I, De Wolf-Peeters C, Falini B, Verhoef G, Morris SW, Hagemeijer A, Van den Berghe H . 1998 Blood 92: 2688–2695
Wong S, Witte ON . 2001 Oncogene 20: 5644–5659
Woolford J, McAuliffe A, Rohrschneider LR . 1988 Cell 55: 965–977
Xu F, Taki T, Yang HW, Hanada R, Hongo T, Ohnishi H, Kobayashi M, Bessho F, Yanagisawa M, Hayashi Y . 1999 Br. J. Haematol. 105: 155–162
Yamamoto Y, Kiyoi H, Nakano Y, Suzuki R, Kodera Y, Miyawaki S, Asou N, Kuriyama K, Yagasaki F, Shimazaki C, Akiyama H, Saito K, Nishimura M, Motoji T, Shinagawa K, Takeshita A, Saito H, Ueda R, Ohno R, Naoe T . 2001 Blood 97: 2434–2439
Yamanashi Y, Baltimore D . 1997 Cell 88: 205–211
Yan XQ, Brady G, Iscove NN . 1993 J. Immunol. 150: 2440–2448
Yang M, Chesterman CN, Chong BH . 1995 Br. J. Haematol. 91: 285–289
Yarden Y, Kuang WJ, Yang-Feng T, Coussens L, Munemitsu S, Dull TJ, Chen E, Schlessinger J, Francke U, Ullrich A . 1987 EMBO J. 6: 3341–3351
Yates KE, Lynch MR, Wong SG, Slamon DJ, Gasson JC . 1995 Oncogene 10: 1239–1242
Yee SP, Mock D, Greer P, Maltby V, Rossant J, Bernstein A, Pawson T . 1989 Mol. Cell. Biol. 9: 5491–5499
Yeung YG, Berg KL, Pixley FJ, Angeletti RH, Stanley ER . 1992 J. Biol. Chem. 267: 23447–23450
Yeung YG, Wang Y, Einstein DB, Lee PS, Stanley ER . 1998 J. Biol. Chem. 273: 17128–17137
Yokote K, Margolis B, Heldin CH, Claesson-Welsh L . 1996 J. Biol. Chem. 271: 30942–30949
Yokote K, Mori S, Hansen K, McGlade J, Pawson T, Heldin CH, Claesson-Welsh L . 1994 J. Biol. Chem. 269: 15337–15343
Yokouchi M, Wakioka T, Sakamoto H, Yasukawa H, Ohtsuka S, Sasaki A, Ohtsubo M, Valius M, Inoue A, Komiya S, Yoshimura A . 1999 Oncogene 18: 759–767
Yu G, Glazer RI . 1987 J. Biol. Chem. 262: 17543–17548
Yu G, Smithgall TE, Glazer RI . 1989 J. Biol. Chem. 264: 10276–10281
Yuan ZM, Huang Y, Ishiko T, Kharbanda S, Weichselbaum R, Kufe D . 1997a Proc. Natl. Acad. Sci. USA 94: 1437–1440
Yuan ZM, Huang Y, Ishiko T, Nakada S, Utsugisawa T, Kharbanda S, Wang R, Sung P, Shinohara A, Weichselbaum R, Kufe D . 1998 J. Biol. Chem. 273: 3799–3802
Yuan ZM, Huang Y, Whang Y, Sawyers C, Weichselbaum R, Kharbanda S, Kufe D . 1996 Nature 382: 272–274
Yuan ZM, Shioya H, Ishiko T, Sun X, Gu J, Huang YY, Lu H, Kharbanda S, Weichselbaum R, Kufe D . 1999 Nature 399: 814–817
Yuan ZM, Utsugisawa T, Huang Y, Ishiko T, Nakada S, Kharbanda S, Weichselbaum R, Kufe D . 1997b J. Biol. Chem. 272: 23485–23488
Zhang Q, Raghunath PN, Xue L, Majewski M, Carpentieri DF, Odum N, Morris S, Skorski T, Wasik MA . 2002 J. Immunol. 168: 466–474
Zhang S, Broxmeyer HE . 1999 Biochem. Biophys. Res. Commun. 254: 440–445
Zhang S, Broxmeyer HE . 2000 Biochem. Biophys. Res. Commun. 277: 195–199
Zhang S, Fukuda S, Lee Y, Hangoc G, Cooper S, Spolski R, Leonard WJ, Broxmeyer HE . 2000 J. Exp. Med. 192: 719–728
Zhang X, Ren R . 1998 Blood 92: 3829–3840
Zhang X, Subrahmanyam R, Wong R, Gross AW, Ren R . 2001a Mol. Cell. Biol. 21: 840–853
Zhang X, Wong R, Hao SX, Pear WS, Ren R . 2001b Blood 97: 277–287
Zirngibl R, Schulze D, Mirski SE, Cole SP, Greer PA . 2001 Exp. Cell. Res. 266: 87–94
Acknowledgements
B Scheijen provided the concept, design, collected the data, drafted the paper and gave final approval. JD Griffin drafted the paper and gave final approval. This work was supported by a fellowship of the Dutch Cancer Society (KWF) to B Scheijen.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Scheijen, B., Griffin, J. Tyrosine kinase oncogenes in normal hematopoiesis and hematological disease. Oncogene 21, 3314–3333 (2002). https://doi.org/10.1038/sj.onc.1205317
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.onc.1205317
Keywords
This article is cited by
-
The telomere complex and the origin of the cancer stem cell
Biomarker Research (2021)
-
Clinicopathologic characterization and abnormal autophagy of CSF1R-related leukoencephalopathy
Translational Neurodegeneration (2019)
-
Hereditary diffuse leukoencephalopathy with axonal spheroids (HDLS): update on molecular genetics
Neurological Sciences (2016)
-
CSF1R mutations in hereditary diffuse leukoencephalopathy with spheroids are loss of function
Scientific Reports (2013)
-
A novel antitumor piperazine alkyl compound causes apoptosis by inducing RhoB expression via ROS-mediated c-Abl/p38 MAPK signaling
Cancer Chemotherapy and Pharmacology (2013)