Abstract
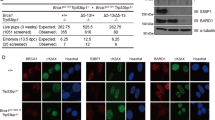
A mouse model with a targeted mutation in the 3′ end of the endogenous Brca1 gene, Brca11700T, was generated to compare the phenotypic consequences of truncated Brca1 proteins with other mutant Brca1 models reported in the literature to date. Mice heterozygous for the Brca11700T mutation do not show any predisposition to tumorigenesis. Treatment of these mice with ionizing radiation or breeding with Apc, Msh-2 or Tp53 mutant mouse models did not show any change in the tumor phenotype. Like other Brca1 mouse models, the Brca11700T mutation is embryonic lethal in homozygous state. However, homozygous Brca11700T embryos reach the headfold stage but are delayed in their development and fail to turn. Thus, in contrast to Brca1null models, the mutant embryos do not undergo growth arrest leading to a developmental block at 6.5 dpc, but continue to proliferate and differentiate until 9.5 dpc. Homozygous embryos die between 9.5–10.5 dpc due to massive apoptosis throughout the embryo. These results indicate that a C-terminal truncating Brca1 mutation removing the last BRCT repeat has a different effect on normal cell function than does the complete absence of Brca1.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Bennett LM, Haugen-Strano A, Cochran C, Brownlee HA, Fiedorek Jr FT, Wiseman RW . 1995 Genomics 29: 576–581
Blaszyk H, Hartmann A, Cunningham JM, Schaid D, Wold LE, Kovach JS, Sommer SS . 2000 Int. J. Cancer 89: 32–38
Bork P, Hofmann K, Bucher P, Neuwald AF, Altschul S, Koonin EV . 1997 FASEB J. 11: 68–76
Chandler J, Hohenstein P, Swing DA, Tessarollo L, Sharan SK . 2001 Genesis 29: 72–77
Chapman M, Verma IM . 1996 Nature 382: 678–679
Chen JJ, Silver DP, Walpita D, Cantor SB, Gazdar AF, Tomlinson G, Couch FJ, Weber BL, Ashley T, Livingston DM, Scully R . 1998 Mol. Cell. 2: 317–328
Fodde R, Edelmann W, Yang K, van Leeuwen C, Carlson C, Renault B, Breukel C, Alt E, Lipkin M, Khan PM, et al. 1994 Proc. Natl. Acad. Sci. USA 91: 8969–8973
Gayther SA, Warren W, Mazoyer S, Russell PA, Harrington PA, Chiano M, Seal S, Hamoudi R, Van Rensburg EJ, Dunning AM, Love R, Evans G, Easton D, Clayton D, Stratton MR, Ponder BAJ . 1995 Nature Genet. 11: 428–433
Gowen LC, Avrutskaya AV, Latour AM, Koller BH, Leadon SA . 1998 Science 281: 1009–1012
Gowen LC, Johnson BL, Latour AM, Sulik KK, Koller BH . 1996 Nature Genet. 12: 191–194
Grade K, Hoffken K, Kath R, Nothnagel A, Bender E, Scherneck S . 1997 J. Cancer Res. Clin. Oncol. 123: 69–70
Hakem R, De la Pompa JL, Sirard C, Mo R, Woo M, Hakem A, Wakeham A, Potter J, Reitmair A, Billia F, Firpo E, Hui CC, Roberts J, Rossant J, Mak TW . 1996 Cell 85: 1009–1023
Hakem R, Delapompa JL, Eli A, Potter J, Mak TW . 1997 Nat. Genet. 16: 298–302
Houven van Oordt CWvd, Smits R, Schouten TG, Houwing-Duistermaat JJ, Williamson SL, Luz A, Meera Khan P, van der Eb AJ, Breuer ML, Fodde R . 1999 Genes Chromosomes Cancer 24: 191–198
Houven van Oordt CWvd, Smits R, Williamson SLH, Luz A, Meera Khan P, Fodde R, Eb AJvd, Breuer ML . 1997 Carcinogenesis 18: 2197–2203
Jacks T, Remington L, Williams BO, Schmitt EM, Halachmi S, Bronson RT, Weinberg RA . 1994 Curr. Biol. 4: 1–7
Koonin E, Altschul S, Bork P . 1996 Nature Genet. 13: 266–268
Liu CY, Fleskennikitin A, Li S, Zeng YY, Lee WH . 1996 Genes Dev. 10: 1835–1843
Ludwig T, Chapman DL, Papaioannou VE, Efstratiadis A . 1997 Genes Dev. 11: 1226–1241
Miki Y, Swensen J, Shattuck-Eidens D, Futreal PA, Harshman K, Tavtigian S, Liu Q, Cochran C, Bennett LM, Ding W, Bell R, Rosenthal J, Hussey C, Tran T, McClure M, Frye C, Hattier T, Phelps R, Haugen-Strano A, Katcher H, Yakumo K, Gholami Z, Shaffer D, Stone S, Bayer S, Wray C, Bogden R, Dayananth P, Ward J, Tonin P, Narod S, Bristow PK, Norris FH, Helvering L, Morrison P, Rosteck P, Lai M, Barret JC, Lewis C, Neuhausen S, Cannon-Albright L, Goldgar D, Wiseman R, Kamb A, Skolnick MH . 1994 Science 266: 66–71
Monteiro AN, August A, Hanafusa H . 1996 Proc. Natl. Acad. Sci. USA 93: 13595–13599
Moynahan ME, Chiu JW, Koller BH, Jasin M . 1999 Mol. Cell 4: 511–518
Ouchi T, Monteiro ANA, August A, Aaronson SA, Hanafusa H . 1998 Proc. Natl. Acad. Sci. USA 95: 2302–2306
Sah V, Attardi LD, Mulligan GJ, Williams BO, Bronson RT, Jacks T . 1995 Nature Genet. 10: 175–180
Scully R, Chen JJ, Ochs RL, Keegan K, Hoekstra M, Feunteun J, Livingston DM . 1997a Cell 90: 425–435
Scully R, Chen JJ, Plug A, Xiao YH, Weaver D, Feunteun J, Ashley T, Livingston DM . 1997b Cell 88: 265–275
Sharan SK, Morimatsu M, Albrecht U, Lim DS, Regel E, Dinh C, Sands A, Eichele G, Hasty P, Bradley A . 1997 Nature 386: 804–810
Shen S-X, Weaver Z, Xu X, Li C, Weinstein M, Chen L, Guan X-Y, Ried T, Deng C-X . 1998 Oncogene 17: 3115–3124
Smits R, Hofland N, Edelmann W, Geugien M, Jagmohan-Changur S, Albuquerque C, Breukel C, Kucherlapati R, Kielman MF, Fodde R . 2000 Genes Chr. Cancer 29: 229–239
Smits R, Kielman MF, Breukel C, Zurcher C, Neufeld K, Jagmohan-Changur S, Hofland N, van Dijk J, White R, Edelmann W, Kucherlapati R, Khan PM, Fodde R . 1999 Genes Dev. 13: 1309–1321
Smits R, van der Houven van Oordt W, Luz A, Zurcher C, Jagmohan-Changur S, Breukel C, Khan PM, Fodde R . 1998 Gastroenterology 114: 275–283
Sobol A, Stoppalyonnet D, Bressacdepaillerets B, Peyrat JP, Kerangueven F, Janin N, Noguchi T, Eisinger F, Guinebretiere JM, Jacquemier J, Birnbaum D . 1996 Cancer Res. 56: 3216–3219
Sobol H, Stoppalyonnet D, Bressacdepaillerets B, Peyrat JP, Guinebretiere JM, Jacquemier J, Eisinger F, Birnbaum D . 1997 Int. J. Oncol. 10: 349–353
Thakur S, Zhang HB, Peng Y, Le H, Carroll B, Ward T, Yao J, Farid LM, Couch FJ, Wilson RB, Weber BL . 1997 Mol. Cell Biol. 17: 444–452
Wang Y, Cortez D, Yazdi P, Neff N, Elledge SJ, Qin J . 2000 Genes Dev. 14: 927–939
Wilkinson DG . 1992 In Situ Hybridization: A Practical Approach. Wilkinson, DG (ed.) IRL Press: Oxford pp 75–83
Wilkinson DG, Bhatt S, Herrmann BG . 1990 Nature 343: 657–659
Wilson CA, Payton MN, Elliott G, Buaas FW, Cajulis EE, Grosshans D, Ramos L, Reese DM, Slamon DJ, Calzone FJ . 1997 Oncogene 14: 1–16
Xu XL, Wagner KU, Larson D, Weaver Z, Li CL, Ried T, Hennighausen L, Wynshawboris A, Deng CX . 1999a Nat. Genet. 22: 37–43
Xu XL, Weaver Z, Linke SP, Li CL, Gotay J, Wang XW, Harris CC, Ried T, Deng CX . 1999b Mol. Cell. 3: 389–395
Yu VP, Koehler M, Steinlein C, Schmid M, Hanakahi LA, van Gool AJ, West SC, Venkitaraman AR . 2000 Genes Dev. 14: 1400–1406
Zhang HB, Somasundaram K, Peng Y, Tian H, Zhang HX, Bi DK, Weber BL, Eldeiry WS . 1998 Oncogene 16: 1713–1721
Zhong Q, Chen CF, Li S, Chen YM, Wang CC, Xiao J, Chen PL, Sharp ZD, Lee WH . 1999 Science 285: 747–750
Acknowledgements
The authors would like to thank Shantie Jagmohan-Changur, Ron Smits, Heleen van der Klift, and Nandy Hofland for technical assistance and critical reading of the manuscript.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Hohenstein, P., Kielman, M., Breukel, C. et al. A targeted mouse Brca1 mutation removing the last BRCT repeat results in apoptosis and embryonic lethality at the headfold stage. Oncogene 20, 2544–2550 (2001). https://doi.org/10.1038/sj.onc.1204363
Received:
Revised:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/sj.onc.1204363
Keywords
This article is cited by
-
BRCA1 and homologous recombination: implications from mouse embryonic development
Cell & Bioscience (2020)
-
Mouse models of BRCA1 and their application to breast cancer research
Cancer and Metastasis Reviews (2013)
-
Deregulation of BRCA1 Leads to Impaired Spatiotemporal Dynamics of γ-H2AX and DNA Damage Responses in Huntington’s Disease
Molecular Neurobiology (2012)
-
Multifunctional transcription factor TFII-I is an activator of BRCA1 function
British Journal of Cancer (2011)
-
Identification of DBC1 as a transcriptional repressor for BRCA1
British Journal of Cancer (2010)