Abstract
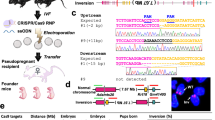
Complex chromosomal rearrangements (deletions, inversions, translocations) are a hallmark of human tumour cells. Yet, the generation of animal models for gross chromosomal abnormalities still presents a formidable challenge. Here, we describe a versatile procedure for chromosomal engineering that was used to generate an ES cell line with a megabase deletion encompassing the tumour suppressor gene neurofibromatosis-1 (Nf-1) on mouse chromosome 11, which is often deleted in tumours of neural crest origin. Homologous recombination into sites flanking Nf-1 was used to introduce artificial sequences (triple-helix, loxP, vector backbone) that can be employed for in vitro recovery of intervening sequences or the generation of in vivo deletions. This strategy may be developed into a scheme by which large chromosomal regions with precisely defined end points may be excised from mammalian cells and reintroduced after suitable in vitro modification.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Gagneten S, Le Y, Miller J and Sauer B. . 1997 Nucl. Acids Res. 25: 3326–3331.
Hochman L, Segev N, Sternberg N and Cohen G. . 1983 Virology 131: 11–17.
Johansson B, Mertens F and Mitelman F. . 1993 Genes Chrom. Cancer 8: 205–218.
Kempkes B, Pich D, Zeidler R and Hammerschmidt W. . 1995 Proc. Natl. Acad. Sci. USA 92: 5875–5879.
Li ZW, Stark G, Gotz J, Rulicke T, Gschwind M, Huber G, Muller U and Weissmann C. . 1996 Proc. Natl. Acad. Sci. USA 93: 6158–6162.
Lichter P, Tang CC, Call K, Hermanson G, Evans GA, Housman D and Ward DC. . 1990 Science 247: 64–69.
Montgomery JC, Silverman KA and Buchberg MA. . 1998 Mammalian Genome 8: 5215–5240.
Nehls M, Krause S and Boehm T. . 1994 Mammalian Genome 5: 183–186.
Nehls M, Kyewski B, Messerle M, Waldschütz R, Schüddekopf K, Smith AJH and Boehm T. . 1996 Science 272: 886–889.
Nehls M, Lüno K, Schorpp M, Pfeifer D, Krause S, Matysiak-Scholze U, Dierbach H and Boehm T. . 1995 Mammalian Genome 6: 321–331.
Ramirez-Solis R, Liu P and Bradley A. . 1995 Nature 378: 720–724.
Sheng Y, Mancino V and Birren B. . 1995 Nucl. Acids Res. 23: 1990–1996.
Shizuya H, Birren B, Kim UJ, Mancino V, Slepak T, Tachiiri Y and Simon M. . 1992 Proc. Natl. Acad. Sci. USA 15: 8794–8797.
Smith AJ, De Sousa MA, Kwabi-Addo B, Heppell-Parton A, Impey H and Rabbitts P. . 1995 Nat. Genet. 9: 376–385.
van Deursen J, Fornerod M, Van Rees B and Grosveld G. . 1995 Proc. Natl. Acad. Sci. USA 92: 7376–7380.
Viskochil D, White R and Cawthon R. . 1993 Annu. Rev. Neurosci. 16: 183–205.
Zhang Y, Buchholz F, Muyrers JP and Stewart AF. . 1998 Nat. Genet. 20: 123–128.
Zhu H and Dean RA. . 1999 Nucl. Acids Res. 27: 910–911.
Acknowledgements
We thank Drs M Nehls and M Messerle for the initial isolation and characterization of genomic clones containing markers D11Bhm100 and D11Bhm109, Dr B Sauer for provision of the GFP-cre expression plasmid, and M Sator-Schmitt and H Kohler for excellent technical assistance. This work was initiated at the German Cancer Research Center, Heidelberg, and completed at the Max-Planck-Institute for Immunobiology, Freiburg. Financial support from the Deutsche Forschungsgemeinschaft is gratefully acknowledged.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Schlake, T., Schupp, I., Kutsche, K. et al. Predetermined chromosomal deletion encompassing the Nf-1 gene. Oncogene 18, 6078–6082 (1999). https://doi.org/10.1038/sj.onc.1203021
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/sj.onc.1203021
Keywords
This article is cited by
-
Engineering chromosomal rearrangements in mice
Nature Reviews Genetics (2001)