Abstract
In familial Beckwith-Wiedemann syndrome (BWS), the mode of inheritance is uncertain and possible patterns include autosomal dominant, multifactorial and autosomal dominant sex-dependent inheritance. Genomic imprinting has recently been invoked to explain the unusual inheritance patterns in several disorders. We have previously reviewed 28 published kindreds of BWS and shown that paternal imprinting is probably responsible for familial BWS. In the present paper, highly informative RFLP markers in the 11p15.5 region have been shown to segregate with the disease gene as an autosomal dominant, but phenotypic manifestations in an offspring are dependent on the sex of the parent contributing the defective gene. In contrast to previous reports in which imprinting of the growth stimulator gene, IGF2, has been invoked as the mechanism explaining sporadic cases of BWS (especially in situations where uniparental disomy and trisomy of the 11p15.5 region has occurred), it is suggested that paternal imprinting of a growth suppressor gene, e.g., H19, may be one of the causes of familial BWS.
Similar content being viewed by others

Introduction
Beckwith-Wiedemann syndrome (BWS) is characterised by variable clinical combinations of gigantism, macroglossia, exomphalos, visceromegaly and ear lobe creases [1,2]. The majority of individuals with BWS reported to date are sporadic, but at least 28 kindreds with familial BWS have been published [3]. There is no clear-cut pattern of inheritance in the familial cases but a key observation towards defining the aetiology of BWS has been the linkage of the disorder to chromosome 1lpl5.5, and a suggestion as to the role of parental imprinting in the transmission of the phenotype [4].
Genomic imprinting is a mechanism which involves a functional difference between maternally and paternally derived genes [5, 6]. Imprinting has been observed in several organisms including the laboratory mouse and it is also evident in some human disorders, including Angelman and Prader-Willi syndromes [6, 7].
We carried out linkage analysis with highly polymorphic 1l p15.5 markers which are known to be tightly linked to the BWS locus in an attempt to decipher the path of the defective gene in a South African BWS family described previously [3]. Our results indicate a clear dominant pattern of inheritance in this unique family with expression of the disorder only when the allele is inherited maternally.
Patients and Methods
BWS was documented in 5 living members of two generations of a South African family of Asian and mixed ancestry. In addition the autopsy evidence confirmed that 2 deceased persons had also been affected. The mode of transmission was unusual, and did not conform to Mendelian patterns of inheritance.
Southern Blots
DNA was prepared from lymphocytes as described previously [8]. 10 µg of DNA were digested with the appropriate restriction endonucleases as recommended by the manufacturer (Boehringer-Mannheim). DNA was electrophoresed and then transferred to nylon membranes (Hybond N+) as described by Southern [9]. Hybridization with the 32P-labelled probes (specific activity, 2 × 108 cpm/µg) was as described previously [10].
Allele Segregation/Linkage Analysis
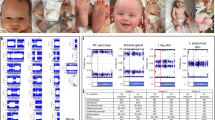
To investigate the passage of the disease locus in the BWS kindred the following closely linked RFLP-identifying probes on chromosome 11p15.5 were used: (a) phins214 (locus INS, ATCC 57398) [11], (b) pbc-N1 (locus HRAS, ATCC 41001) [12]. The genotyped pedigree of the kindred investigated in this study is shown in figure 1; the clinical details have been reported elsewhere [3, 13]. Linkage analysis was carried out with the program LINKAGE (V5.03) [14]. Previously estimated values for incidence (1:13,500) and population disease frequency (0.0001) [4] were used. A value of 10−5 was assumed for the spontaneous mutation rate. Two liability classes of penetrance were assigned to members of the family according to the model for paternal imprinting as proposed previously [3]. For class 1 the penetrance was 0.0 for the disease gene heterozygote and homozygote and for liability class 2 a penetrance value of 1.0 was assigned for both homozygote and heterozygote. Class 1 comprised of individuals from generations I and II and in marrying spouses; class 2 consisted of family members who were descendents of II-1/II-2, II-8/II-9, and III-3/III-4.
Results
In the South African BWS kindred (fig. 1) in which seven individuals have BWS, two phenotypically normal half-sisters (II-2, II-8) who have a common father (I-2), each produced three offspring (III-2, III-4, III-5 and III-14, III-16, III-19) with the characteristic features of BWS. Two affected boys (III-2, III-5) died in early infancy with conclusive autopsy evidence of the disorder. The remaining four living relatives are all female, the eldest of whom (III-4) has had a baby with BWS (IV-1). Although a clearly autosomal dominant mode of inheritance was not evident from the pedigree of the BWS family, linkage analysis was carried out assuming this mode of inheritance with the two liabilty classes of penetrance assigned to members of the family according to the model for paternal imprinting as proposed previously [3]. Generations I and II were accorded a penetrance of 0.0 because of the parental origin of the defective allele. Individual I-2 was an obligate gene carrier by virtue of pedigree data but as he did not manifest any stigmata of BWS, he was accorded a penetrance of 0.0.
RFLP studies were carried out solely for the purpose of tracking marker alleles in relation to the BWS phenotype and not to establish linkage (which has previously been shown [4]). The genotypes for phins214 are shown on the pedigree in fig. 1. Tight linkage of BWS to the 11p15.5 region was confirmed by a maximum LOD score of 2.74 at a recombination frequency of 0.00 (table 1).
Discussion
Normal growth and development in organisms is due to a fine balance between the activities of growth enhancers and suppressors. It is likely that the BWS phenotype is the result of one or a combination of several mechanisms affecting the functioning of either a growth stimulant or a growth suppressor. A defective or absent growth suppressor is likely to result in generalized overgrowth. On the other hand any duplication of the functional gene for a growth enhancer is also likely to produce generalized overgrowth.
A model involving maternal imprinting has been proposed by Henry et al. [15] to explain observations of uniparental paternal disomy and 11p15.5 trisomy (with paternal disomy) in sporadic BWS patients. A growth-promoting substance is presumed to be coded for by that BWS gene which is maternally imprinted. In the model of Henry et al. [15], paternal disomy would effectively result in a double dose of the active paternal growth factor gene product resulting in phenotypic overgrowth, typical of BWS. In this regard the insulin-like growth factor gene (IGF2) has been implicated as the candidate for BWS [16]. The IGF2 gene located at 11p15.5 on the human genome occurs on a syntenically homologous region of the mouse genome and has been observed to be maternally imprinted in laboratory mice [17, 18].
We have reviewed the literature and, in contrast to the above model, showed that paternal imprinting is likely to account for the abnormal pattern of inheritance in 26 of the 29 previously reported BWS kindreds [3]. Our data supported the theory of maternal transmission in familial BWS suggested by Koufos et al. [4]. In the present article, using informative RFLP markers to show the passage of the BWS allele in the South African family, we further argue in favour of paternal imprinting of a growth suppressor as a cause of familial BWS. The two RFLP markers used (phins214 and pbc-N1) showed no recombination with the disease locus. However, only results for the more informative probe, phins214, are shown in figure 1 and will be discussed here. As shown in figure 1, a single allele (B) segregates from the carrier, I-2, to each of the affected individuals through a non-manifesting parent. This apparent lack of penetrance in the obligate carriers in generation II (II-2 and II-8) is consistent with our theory that paternal imprinting of the defective (B) BWS allele results in no apparent effect on the phenotype of carriers in generation II.
The postulated growth suppressor, also located in the 11p15.5 region, would always be functional only on the maternal gene, and would be required in a single dose in order to regulate cell growth. The paternally derived allele would be expected to be non-functional. In this way a defect in the usually active maternally derived allele would give rise to the production of a defective or non-functional growth suppressor, resulting in a phenotype of generalized overgrowth, as in BWS.
Our model is also consistent with the observation of Henry et al. [15] of uniparental paternal disomy in sporadic BWS patients, since any allele at the proposed ‘growth repressor’ BWS locus of paternal origin is imprinted. This factor would result in lack of suppressor function in paternal disomics (whether isodisomic or heterodisomic) and an affected individual would have features of generalised overgrowth. In addition to the lack of growth suppressor in individuals paternally disomic for 11p15.5, the IGF2 gene would be present in a paternally active double dose, as proposed by Henry et al. [15].
The model proposed here also accounts for the apparent higher penetrance in individuals born to female carriers, as is evident in the family described in this study. Although decreased fecundity in transmitting males is a factor, as demonstrated by Moutou et al. [19], the reported excess of female carriers in BWS is very likely to be due to the expression of the defective BWS allele only in offspring inheriting the allele from their mothers; inheritance of a paternally derived defective gene (which is imprinted, anyway) in offspring will not produce any phenotypic effect. Therefore, as with individual I-2 (and II-11) in the reported family, a non-manifesting carrier male could only be identified as such through the offspring of his (phenotypically normal carrier) daughters.
Although we have not addressed the issue of BWS-associated tumors, the observation of consistent maternal allele loss in tumors associated with BWS may support the hypothesis of a normal functional maternal allele being required for a normal phenotype.
Rather than BWS being due to the effect exclusively of one gene, e.g., a growth factor as suggested previously, a likely scenario is that two closely located genes are involved in the aetiology of BWS as suggested by Little et al. [16], one coding for a growth suppressor, which is paternally imprinted and another coding for a growth stimulator, which is maternally imprinted. Two such candidate genes are the insulin-like growth factor 2 (IGF2) gene, which is a paternally active growth enhancer gene, and nearby, a recently located maternally active growth suppressor gene, H19 [20]. Reciprocal imprinting of different parental alleles of one or other of these two loci would explain the apparent paradoxical mechanisms of maternal imprinting in sporadic BWS and the converse in familial cases of BWS. It has been shown that mice embryos paternally disomic for the chromosomal region homologous to human 11p15.5 exhibit overgrowth, and those that are maternally disomic for this region are growth retarded [18].
In summary, our linkage data and review of familial cases strongly suggest paternal imprinting in BWS. Acceptance of this hypothesis allows clear genetic counselling for individuals in some BWS families. In addition, the highly polymorphic loci which are very tightly linked to the BWS locus make possible precise antenatal diagnosis of pregnancies in clinically normal female carriers.
References
Wiedemann HR: Complexe malfor-matif familial avec hernie ombilicale et macroglossie — un «Syndrome nouveau»? J Génét Hum 1964;13:223–232
Beckwith JB: Macroglossia, omphalocele, adrenal cytomegaly, gigantism, and hyperplastic visceromegaly. Birth Defects 1969;2:188–196
Viljoen DL, Ramesar RS: Evidence for paternal imprinting in familial Beckwith-Wiedemann Syndrome. J Med Genet 1992;29:221–225
Koufos A, Grundy P, Morgan K, Aleck KA, Hadro T, Lampkin BC, Kalbakji A, Cavanee WK: Familial Wiedemann-Beckwith syndrome and a second Wilms tumor locus both map to 11p15.5. Am J Hum Genet 1989;44:711–719
Hall JG: How imprinting is relevant to human disease. Development 1990;(suppl):141–148.
Sapienza C: Parental imprinting of genes. Sci Am 1990;263:26–32
Clarke A: Genetic imprinting in clinical genetics. Development 1990;(suppl):131–139.
Van Den Plas S, Wiid I, Grobler-Rabie A, Brebner K, Ricketts M, Wallis G, Bester A: Blot hybridization analysis of genomic DNA. J Med Genet 1984;21:164–172
Southern EM: Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol 1975;98:5503–5517
Feinberg AP, Vogelstein B: A technique for the radiolabelling of DNA of restriction fragments to a high specific activity. Anal Biochem 1984;137:266–267
Bell G, Horita S, Karam JH: A polymorphic locus near the human insulin gene is associated with insulin-dependent diabetes mellitus. Diabetes 1984;3:176–183
Pierotti MA; Radice P, Biunno I, Borrello MG, Cattadori MR, Porta GD: Two RFLPs generated by TaqI at the human HRAS1 locus. Nucleic Acids Res 1986;14:4379.
Viljoen DL, Jacquire Z, Woods DL: Prenatal diagnosis in autosomal dominant Beckwith-Wiedemann syndrome. Prenat Diagn 1991;11:167–175
Lathrop GM, Laouel JM: Easy calculations of LOD scores and genetic risks on small computers. Am J Hum Genet 1984;36:460–465
Henry I, Jean-Pierre M, Barichard F, Serre JL; Mallet J, Turleau C, de Grouchy J, Junien C: Duplication of HRAS1, INS, and IGF2 is not a common event in Beckwith-Wiedemann syndrome. Ann Genet 1988;31:216–220
Little M, Van Heyningen V, Hastie N: Dads, disomy and disease. Nature 1991;351:609–610
DeChiara TM, Robertson EJ, Efstratiadis A: Parental imprinting of the mouse insulin-like growth factor II gene. Cell 1991;64:849–859
Ferguson-Smith AC, Cattanach BM, Barton SC, Boechey CV, Surani MA. Embryological and molecular investigations of parental imprinting on mouse chromosome 7. Nature 1991;351:667–670.
Moutou C, Junien C, Henry I, Bonaiti-Pellie: Beckwith-Wiedemann syndrome: A demonstration of the mechanisms responsible for the excess of transmitting females. J Med Genet 1992;29:217–220
Bartolomei MS; Zemel S, Tilghman SM: Parental imprinting of the mouse H19 gene. Nature 1991;351:153–155
Acknowledgements
This research was supported by grants from the South African Medical Research Council, the Mauer-berger Fund, the Harry Crossley Foundation and the University of Cape Town Staff Research Fund.
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Ramesar, R., Babaya, M. & Viljoen, D. Molecular Investigation of Familial Beckwith-Wiedemann Syndrome: A Model for Paternal Imprinting. Eur J Hum Genet 1, 109–113 (1993). https://doi.org/10.1159/000472397
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1159/000472397