Abstract
A key mechanism employed by plants to adapt to salinity stress involves maintaining ion homeostasis via the actions of ion transporters. While the function of cation transporters in maintaining ion homeostasis in plants has been extensively studied, little is known about the roles of their anion counterparts in this process. Here, we describe a mechanism of salt adaptation in plants. We characterized the chloride channel (CLC) gene AtCLCf, whose expression is regulated by WRKY transcription factor under salt stress in Arabidopsis thaliana. Loss-of-function atclcf seedlings show increased sensitivity to salt, whereas AtCLCf overexpression confers enhanced resistance to salt stress. Salt stress induces the translocation of GFP-AtCLCf fusion protein to the plasma membrane (PM). Blocking AtCLCf translocation using the exocytosis inhibitor brefeldin-A or mutating the small GTPase gene AtRABA1b/BEX5 (RAS GENES FROM RAT BRAINA1b homolog) increases salt sensitivity in plants. Electrophysiology and liposome-based assays confirm the Cl−/H+ antiport function of AtCLCf. Therefore, we have uncovered a mechanism of plant adaptation to salt stress involving the NaCl-induced translocation of AtCLCf to the PM, thus facilitating Cl− removal at the roots, and increasing the plant’s salinity tolerance.
Similar content being viewed by others
Introduction
Increased soil salinity is a growing challenge for crop production, which adversely affects crop yields in ~20% of cultivable land worldwide. This problem is predicted to affect >40% of agricultural lands in the coming decades1. The excess accumulation of soluble salts, especially sodium chloride (NaCl), in the root zone severely impedes plant growth, reducing crop productivity. Previous efforts to identify solutions for mitigating salinity stress have focused primarily on cations such as sodium (Na+)2. It is important to explore the effects of counter-anions such as chloride (Cl−) on root growth to help plants cope with increased soil salinity. Although Cl− is an essential nutrient for crop plants and is used as osmoticum in halophytes3,4, excess Cl− inhibits plant growth and yield5. Cl− efflux from roots is positively correlated with salt tolerance6, suggesting that it might be the key mechanism preventing Cl− toxicity in plants. However, little is known about how roots carry out Cl− efflux to overcome salinity stress.
Recent studies have implicated plant chloride channels (CLCs) as involved in regulating Cl− homeostasis under salinity stress7,8. CLCs play vital roles in stabilizing membrane potential and turgor pressure. These integral membrane proteins contain 10–12 transmembrane domains and are present in all kingdoms of life9,10. Although the functions of plant CLCs are not well-studied, extensive information exists for their animal counterparts, revealing important effects in both normal development and disease states. For example, mutations in mammalian CLC genes cause genetic diseases such as Bartter syndrome, idiopathic epilepsy, osteopetrosis, Dent’s disease, and various types of myopathies11,12. However, the effects of CLC mutations in plants are unclear.
Despite their nomenclature, CLC family members are not exclusively Cl− transporters. Although some CLCs function as Cl− channels or chloride-proton antiporters (Cl−/H+), others act as NO3− transporters10,13. The transmembrane regions of Cl−/H+ antiporter CLCs are characterized by two key conserved ‘gating’ and ‘proton’ glutamate residues, whereas the Cl− channels contain only the gating glutamate in the selectivity filter domain9,14. Among the seven CLC genes in Arabidopsis thaliana, we determined that AtCLCf is induced in roots in response to salt treatment. AtCLCf was predicted to be a Cl− channel localized to the Golgi apparatus and the trans-Golgi network/early endosome (TGN/EE)15,16, but how it helps reduce ion toxicity in the roots of plants under salt stress remains unknown. In addition to their function, molecular regulatory mechanism behind the expression of CLCs and the study on associated TFs are limited.
Here, we describe the mode of action of AtCLCf in the remediation of Cl− toxicity under salt stress. Our findings show that AtCLCf expression is directly regulated by WRKY9 transcription factor and the increased intracellular NaCl levels induce the translocation of AtCLCf from the Golgi apparatus to the plasma membrane (PM) via TGN using an AtRABA1b (BEX5)-mediated pathway. The electrophysiological studies (patch clamp) with Human Embryonic Kidney 293 (HEK293FT) cells show that AtCLCf functions as a Cl−/H+ antiporter involved in efflux of Cl− from the cells. The subcellular translocation of this CLC in the root cortex tissue and epidermis represents an essential salt tolerance mechanism in Arabidopsis.
Results
AtCLCf is essential for salt tolerance
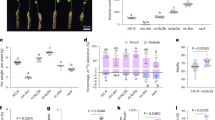
Because roots are the primary plant organs that perceive salt stress, we examined the role of AtCLCf in root elongation using one-week-old Arabidopsis seedlings of different genotypes. The roots of atclcf seedlings were sensitive to 6 h of 50 mM NaCl treatment, showing significant reductions in the lengths of the meristematic zone (MZ, 20%) and elongation zone (EZ, 44%) (Fig. 1a–f) that led to shorter roots (Fig. 1g). By contrast, complemented atclcf (pAtCLCf::AtCLCf;atclcf) plants exhibited phenotypic rescue, as they grew in a similar manner to the wild type (WT) under salt stress (Fig. 1a–g), with significantly lower NaCl-induced reductions in root growth occurring in both pAtCLCf::AtCLCf;atclcf and 35S::AtCLCf;atclcf plants compared to atclcf. Additionally, atclcf seedling root elongation was inhibited by KCl treatment, but not by NaNO3 or the osmotic stress from mannitol treatment (Supplementary Fig. 1), suggesting that the effects seen are specific to Cl− stress.
DIC microscopy images of the root tips of one week-old seedlings of different genotypes grown (a) without and (b) with salt treatment (50 mM NaCl, 6 h). Arrows indicate the border between the meristematic zone (MZ) and elongation zone (EZ). Line indicates the end of the EZ near the root hair formation zone. Scale bar, 200 µm. Root lengths of seedlings (genotypes indicated at the top) grown without (c) and with (d) 50 mM NaCl treatment, at 7 days after germination; scale bar, 1 cm. Root tips are marked by white dots. Quantification of MZ (e) and EZ (f) length from a and b using ImageJ software (mean ± SE, n = 10 per genotype). g Root lengths of seedlings in c and d, mean ± SE, n = 40 per genotype. Means with different letters within a data set (in e–g) are significantly different, P < 0.05 (one-way ANOVA followed by Tukey’s test). Data sets without significant differences are unlabeled. Data were obtained from three independent experiments, and representative images are shown here.
We analyzed the salt responsiveness of AtCLCf transcripts by performing qRT-PCR and GUS reporter assays. In the absence of NaCl treatment, roots and siliques exhibited high AtCLCf transcript levels (Fig. 2a–f). After 6 h of NaCl treatment, AtCLCf transcript levels in the roots increased ~3-fold (Fig. 2g–i and Supplementary Fig. 2a), but those in the shoots did not change significantly. These results suggest that AtCLCf plays an important role in maintaining Cl− homeostasis in the roots. Older Arabidopsis plants showed a similar response to salt (Fig. 2j, k): when treated with NaCl for one week, 35S::AtCLCf;atclcf plants showed significantly higher chlorophyll content (~6 times), fresh weight (Supplementary Table 1 ~1.5 times), and survival rate (~2 times) than atclcf plants (Supplementary Fig. 2b, c). Severe chlorosis and stunted growth were observed in atclcf and WT plants in response to NaCl treatment, and these plants could not recover to the same extent as 35S::AtCLCf;atclcf plants one week after the salt stress was withdrawn (Supplementary Fig. 3a–c). Although salt treatment increased the Cl− contents in the shoots and roots of plants of all genotypes, the effect was greatest in atclcf shoots (Fig. 2l). These results suggest that AtCLCf functions to minimize the amount of Cl− reaching the shoots, likely by removing the excess Cl− at the roots, thereby protecting the photosynthetically active tissues of Arabidopsis from salinity stress.
a Tissue-specific expression of AtCLCf n = 3 (3 biological replicates, each with 3 technical replicates). Data are mean ± SE, means with different letters within a data set are significantly different, P < 0.05 (one-way ANOVA followed by Tukey’s test). pAtCLCf::GUS expression in one-month-old Arabidopsis (b) root, (c) flower, (d) leaf, (e) silique, and (f) leaf vasculature. Scale bar in b and f = 100 µm, c–e = 500 µm, data were obtained from three independent experiments, and representative images are shown in b–f. g Time-course analysis of AtCLCf expression in the shoots and roots of one-week-old Arabidopsis seedlings after 100 mM NaCl treatment. Data are mean ± SE, n = 3 (3 biological replicates, each with 3 technical replicates). Asterisks indicate statistically significant differences (*P < 0.05, **P < 0.01) between 0 h and other time points, as measured by unpaired Student’s t test (two-tailed). h pAtCLCf::GUS expression in one-week-old untreated and salt-treated (100 mM NaCl, 6 h) Arabidopsis seedlings. Scale bar = 1 mm, data were obtained from three independent experiments, and representative images are shown here. i Close-up images of the roots in h Scale bar = 100 µm. j, k Growth responses of one-month-old WT, atclcf, and 35S::AtCLCf plants grown in soil (j) without NaCl treatment and (k) after one week of 150 mM NaCl treatment. Scale bar = 1 cm, (n = 3). l Cl− contents in the shoots and roots of WT, atclcf, and 35S::AtCLCf plants under control (untreated) and salt-treated conditions (n = 5). Data are mean ± SE, means with different letters within a data set are significantly different, P < 0.05 (one-way ANOVA followed by Tukey’s test). The box plots with whiskers show all the data points and their distribution. Horizontal lines within the boxes indicate the median. Minimal and maximal data points are indicated by the lower and the upper whiskers, respectively, untr untreated, sh shoot, R root. m ChIP-qPCR analysis shows enrichment of AtCLCf promoter fragment in WRKY9 ChIP sample. Data are mean ± SE, n = 3 (3 biological replicates, each with 3 technical replicates). n Luciferase assay carried out using the mesophyll protoplasts obtained from leaves of 4-week-old atwrky9 mutants. pAtCLCf::LUC was used as the control and 35S::AtWRKY9 was used as the test. The AtCLCf promoter fragments with mutated WRKY binding sites were used as additional controls. Firefly luciferase activity was normalized to Renilla luciferase activity and plotted (Mean ± SE, n = 5). Asterisks in m and n indicate statistically significant differences (*P < 0.05, **P < 0.01) between control and test, as measured by Student’s t test.
Perception of salinity signal by AtWRKY9 induces AtCLCf expression
We sought to examine how the salinity signal is transmitted to AtCLCf. Accordingly, we analyzed the 5’-upstream region of AtCLCf and detected abiotic stress-related cis-elements, including WRKY binding domains (Supplementary Fig. 4a). The role of WRKY TFs in perceiving salt is well-established in plants. While ectopic expression of wheat WRKYs increased salt tolerance in Arabidopsis17, expression of several WRKYs such as WRKY9, 25, 33 was positively correlated with improved salt tolerance in other species18,19. Also, AoWRKY9 was induced upon salt stress in our earlier study20. Interestingly, AtCLCf expression was significantly suppressed (~10-fold) in atwrky9, but not in atwrky6 and atwrky33 mutants (Supplementary Fig. 4b). Furthermore, ChIP-qPCR showed ~4-fold enrichment of AtCLCf promoter fragment by AtWRKY9-HA, while there was no significant enrichment by the other two WRKYs studied (Fig. 2m). This was supported by independent luciferase assays where ~4-fold higher luminescence was observed in atwrky9 protoplasts transfected with pAtCLCf::LUC along with 35S::AtWRKY9, compared to the control. There was no enhancement in luminescence when the WRKY binding domains in AtCLCf promoter fragment were mutated (Fig. 2n). The interaction between AtWRKY9 and AtCLCf promoter was also confirmed through Y1H assay (Supplementary Fig. 4c). Collectively, these results show that under salt stress conditions AtWRKY9 binds to AtCLCf promoter and enhances its expression.
AtCLCf functions as a Cl−/H+ antiporter and is involved in Cl− efflux
Some CLCs do not transport Cl−15. Hence, we tested whether AtCLCf is a functional Cl− channel using liposomes incorporated with recombinant AtCLCf protein and patch clamp experiments by expressing the AtCLCf in HEK293FT cells (Fig. 3). In addition, vesicles prepared from PMs extracted from WT and atclcf plants treated for 6 h with NaCl were used to check the Cl− transport (Supplementary Fig. 5). Channel activity was quantified as the decrease in fluorescence of a fluorophore (MQAE, loaded into the liposomes or the PM vesicles) that is quenched by Cl− ions. The liposome-based fluorescence quenching assays helped to determine the transport activity of recombinant AtCLCf protein with NaCl, KCl and CaCl2 as Cl− donors. Data from liposomes with the incorporation of recombinant AtCLCf protein showed that the Cl− transport rate was in the following order: CaCl2 > NaCl > KCl, while the control liposomes without AtCLCf showed no fluorescence quenching (Fig. 3a). Increasing the concentration of NaCl in the external buffer showed a concentration dependent increase in fluorescence quenching (Fig. 3b), confirming that AtCLCf is a functional Cl− channel. Similarly, the patch clamp recordings showed that, by pulsing the cells to different voltages ranging from −80 to 80 mV, AtCLCf transfected HEK293FT cells displayed significantly larger inward and outward current density as compared to vector transfected cells. (Fig. 3). The current density (pA/pF) was scored by normalizing current over cell capacitance of individual cells. The data show a larger inward current (−30.86 ± 6.34) than outward current (23.45 ± 4.54) indicating a stronger outflux of chloride ions (Fig. 3E–G). Subsequently, to test how AtCLCf functions, we recorded the currents by changing the internal pH from 7.4 to 5.5 and found an increase in the influx of anion (Cl−) suggesting an efflux of the H+ ions (Supplementary Fig. 6a–c). Further, its ion selectivity was tested by checking the effect of replacing most of internal CsCl with CsF, CsBr, CsI, CsNO3 or CsAcetate on the ionic current. Interestingly, there was a significant increase in Cl− efflux among the anions tested in AtCLCf transfected cells compared to the vector (Fig. 3h–j). Concomitantly, using the pH-sensitive fluorescent dye HPTS (8-hydroxy-1,3,6-pyrenetrisulfonate) in liposome assays (Supplementary Fig. 6d, e), we observed a significant H+ transport in AtCLCf-embedded liposomes when the external buffer pH was increased to 8.0 and the extravesicular halide anions were changed. Significant changes in the ion transport rate were observed, with the selectivity as: Cl− > F− > I−. These results suggest that the Cl− transport activity of AtCLCf is coupled with H+ exchange. Additionally, to examine the directionality of Cl− transport, we prepared right-side-out (RSO) and inside-out (ISO) PM vesicles (Supplementary Fig. 5) from WT and atclcf plants and used them for the fluorescence quenching assay. No significant Cl− transport was observed in the RSO vesicles (Supplementary Fig. 5a). More importantly, WT ISO vesicles showed higher fluorescence quenching than ISO vesicles from atclcf plants (Supplementary Fig. 5b), suggesting that AtCLCf may have a directional Cl− transport activity from within the cell to the outside across the PM. All these results confirm that AtCLCf functions as a Cl−/H+ antiporter and may be involved in the efflux of Cl− in plant cells.
a Chloride-selective ion transport by MQAE fluorescence quenching assay showed transport of chloride ion by AtCLCf. Data are mean ± SD (n = 3). (Inset to a) Schematic representation of chloride-selective, MQAE dye-based fluorescence assay for ion transport. Liposomes without incorporation of AtCLCf were used as control. Arrow indicates the time point where different salts were added. The schematic was created using PowerPoint software. b AtCLCf showed increased transport of chloride with addition of increasing NaCl concentrations in MQAE based quenching assay. Data are mean ± SD (n = 3). c–e Enhanced inward and outward current was observed in cells expressing AtCLCf as compared to the vector transfected cells. c Exemplar traces of a vector transfected cell showing the current evoked from holding potential of 0 mV to different test potential ranging from −80 mV to 80 mV with 10 mV increment. d Exemplar trace of AtCLCf cell. e Current voltage relationship of respective groups. f, g Data comparing the maximal outward and inward current at 80 and −80 mV respectively. **P < 0.01 (e–g data are mean ± SE, vector n = 12, AtClCf n = 14). Statistics was performed with unpaired Student’s t test (2-tailed). h–j The effect of replacing most of internal CsCl with CsF, CsBr, CsI, CsNO3 or CsAcetate on the ionic current. h Current voltage relationship of AtCLCf transfected cell recorded with 140 mM CsCl-based internal as compared to cell recorded with 130 mM CsF, CsBr, CsI, CsNO3 or CsAcetate and 10 mM CsCl-based internal solution. The profiles for vector and AtCLCf cell recorded with CsCl based internal solution were reproduced from e for comparison, i and j data comparing the maximal outward and inward current at 80 and −80 mV respectively. (h–j data are mean ± SE, n = 12 for CsF, 11 for CsBr, 9 for CsI, 12 for CsNO3 and 9 for CsAcetate). Different letters within a data set are significantly different, P < 0.01 (one-way ANOVA followed by Tukey’s test).
To gain structural insights into the roles of specific amino acid residues in AtCLCf function, we generated a homology model (Fig. 4a–c) based on the crystal structure of Salmonella typhimurium CLC9, which shares 48% sequence similarity with AtCLCf. Three selectivity filter domains were identified in AtCLCf. We mutated five conserved amino acid residues in these domains and studied their effects on AtCLCf function using yeast growth assays (Fig. 4b–d). Specifically, we assayed the Cl− transport function of AtCLCf by introducing the WT AtCLCf gene and AtCLCf genes with mutations in the three selectivity filter domains (domain 1, Δgef1_MD1; domain 2, Δgef1_MD2; and domain 3, Δgef1_MD3) into the yeast (Saccharomyces cerevisiae) deletion mutant Δgef1, which lacks the GEF1 gene (a yeast homolog of CLC genes). The salt sensitivity of Δgef1 mutant strain was rescued by the introduced WT AtCLCf gene and MD1, while introduction of the other two domain mutants (MD2 and MD3) failed to rescue the salt sensitivity (Fig. 4e).
a Homology model of AtCLCf based on the crystal structure of Salmonella typhimurium CLC was generated using PyMOL software and it shows three selectivity filter domains. b Enlarged view of (a) showing the selectivity filter domains and the pore. c, d Positions of mutations in the selectivity filter domains: MD1(I204F, Q207Y); MD2(E247W); MD3(L463W, P466F). e Growth of yeast strain BY4741 and its derivative Δgef1 harboring mutated domains (MD1, MD2, and MD3) and AtCLCf in the YEp352 plasmid grown on SD/-Ura medium with (0.5–1.25 M) and without NaCl, data were obtained from three independent experiments, and representative images are shown here.
To confirm the Cl− transport function of AtCLCf in plant cells, we introduced the constructs harboring mutated AtCLCf and 35S::AtCLCf into atclcf plants and measured the root length of the transgenic seedlings. There was no significant difference in root length between these genotypes in the absence of salt treatment (Fig. 5a). Upon NaCl treatment, 35S::AtCLCf;atclcf plants exhibited higher salt tolerance than atclcf plants, with their root length restored to WT levels. By contrast, transgenic plants harboring mutated 35S::MD2;atclcf (E247W) and 35S::MD3;atclcf (L463W and P466F) behaved similar to atclcf seedlings, indicating that the expression of these constructs failed to rescue the increased salt sensitivity of the mutant (Fig. 5b, c). These results show that the residue that is altered in the 35S::MD2;atclcf mutant, namely, glutamate 247 in the selectivity filter domain 2, is important for the functioning of AtCLCf, as has also been found in other organisms9,10.
Root lengths of WT, atclcf, 35S::AtCLCf;atclcf, and 35S::MD1-MD3AtCLCf;atclcf seedlings, under (a) untreated and (b) treated with 50 mM NaCl conditions; scale bar, 1 cm. Root tips are marked by white dots. c Root lengths of seedlings of the indicated genotypes with (50 mM) and without NaCl treatment. Root lengths were measured after one week of germination, Mean ± SE, n = 20. d Results of Cl− transport assay using mesophyll protoplasts of one-month-old WT, atclcf, 35S::AtCLCf;atclcf, and 35S::MD2AtCLCf;atclcf (35S::MD2) plants. Protoplasts treated with 50 mM NaCl for 3 h were loaded with 100 µM MQAE for 30 min and visualized under a confocal microscope. Lower panel shows the respective brightfield images; scale bars, 20 µm. e Quantification of the data in d. Mean ± SE, n = 10. f Cl− efflux assay (as described in the methods) was carried out from 10-day-old seedlings of WT, atclcf and 35S::AtCLCf;atclcf (n = 5). Means with different letters within a data set are significantly different, P < 0.05 (one-way ANOVA followed by Tukey’s test). Data were obtained from three independent experiments, and representative images are shown here. Horizontal lines within the boxes in e and f indicate the median. Minimal and maximal data points are indicated by the lower and the upper whiskers, respectively.
Having demonstrated that AtCLCf is a functional Cl−/H+ antiporter containing the typical glutamate in its selectivity filter domain, we performed protoplast-based assays to attempt to verify the directionality of transport in the plant. An MQAE assay using mesophyll protoplasts showed that more Cl− was retained in atclcf and 35S::MD2;atclcf protoplasts (exhibit low fluorescence = more quenching) than in 35S::AtCLCf;atclcf and WT protoplasts (Fig. 5d, e). In an independent assay, we quantified the fluorescence quenching (via spectrofluorimetry) of salt-treated protoplasts loaded with MQAE. Similar to the above observations (microscopy), the atclcf and 35S::MD2;atclcf protoplasts showed significantly higher quenching, which shows that more Cl− is retained in them as a result of reduced Cl− efflux compared to the 35S::AtCLCf;atclcf and WT protoplasts (Supplementary Fig. 7). Additionally, there was a significant reduction in Cl− efflux from atclcf seedling roots compared to those of the WT and 35S::AtCLCf;atclcf (Fig. 5f). Collectively, these findings show that AtCLCf is important for removal of excess Cl− from the roots.
Salt stress triggers AtCLCf translocation from the Golgi apparatus to the PM
The earlier observation that AtCLCf localizes to the Golgi apparatus or TGN/EE15,16,21 does not adequately explain how this protein confers salt tolerance at either the cellular or plant level. To address this issue, we first examined the protein’s subcellular localization by transiently expressing 35S::GFP-AtCLCf in Nicotiana benthamiana leaf epidermal cells. In untreated cells, GFP-tagged AtCLCf localized to the Golgi along with the Golgi marker G-rk CD3-967 (Fig. 6a). Interestingly, after 4 h of NaCl treatment, GFP-AtCLCf progressively spread via the TGN (along with the TGN marker SYP61-RFP) to the PM (Fig. 6b). After 6 h of NaCl treatment, the GFP-AtCLCf signal was mostly concentrated in the PM (Fig. 6c and Supplementary Video 1) along with the PM marker (PM-rk CD3-1007). There was about 60% increase in the translocation of AtCLCf to the PM compared to the 0 h value (Fig. 6d), indicating that NaCl induced this subcellular translocation. A similar stimulatory effect of NaCl on the formation of TGVs and their fusion with vacuoles/PM was previously demonstrated for FLOTILLIN1 in Arabidopsis22. We further confirmed the NaCl-induced translocation of AtCLCf to the PM in Arabidopsis leaf protoplasts expressing AtCLCf under its native promoter (pAtCLCf::GFP-AtCLCf) (Fig. 6e). These data suggest that the underlying translocation mechanism involves the exocytic trafficking pathway. We also detected AtCLCf expression in the root cortex and epidermis of pAtCLCf::GFP-AtCLCf seedlings within 6 h of salt treatment (Supplementary Fig. 8a, b). Notably, the differences in the root elongation responses of the lines were observed within the same timeframe of a 6 h NaCl treatment (Fig. 1a, b). Thus, we identified a salt-induced translocation of AtCLCf to the PM, providing a rapid mechanism to help Arabidopsis root cells adapt to salt stress.
a–c Confocal microscopy images of Nicotiana benthamiana leaf epidermal cells show that N-terminal GFP-tagged AtCLCf translocates to the PM when treated with 100 mM NaCl, as indicated by the expression of GFP-AtCLCf along with Golgi, TGN, and PM markers. Shown are GFP-AtCLCf expression (a) before and after (b) 4 h and (c) 6 h of NaCl treatment. The GFP signal was visualized under 488 nm excitation, 500–525 nm emission; the markers were visualized under 561 nm excitation, 575–600 nm emission. Co-localization of GFP-AtCLCf with markers is shown in the merged images. Scale bars, 20 µm, data were obtained from three independent experiments, and representative images are shown here. d Quantification of AtCLCf-GFP localization with respective markers (n = 7) using ImageJ. Horizontal lines within the boxes in d indicate the median. Minimal and maximal data points are indicated by the lower and the upper whiskers, respectively. e Expression of GFP-tagged AtCLCf in the leaf mesophyll protoplasts of pAtCLCf::GFP-AtCLCf Arabidopsis lines without and with NaCl treatment, n = 10. Scale bars, 10 µm. Means with different letters are significantly different, P < 0.05 (one-way ANOVA followed by Tukey’s test).
AtRABA1b/BEX5 mediates the salt-induced translocation of AtCLCf from the Golgi to the PM
Similar to what we observed in N. benthamiana and Arabidopsis (leaves and protoplasts), GFP-AtCLCf translocated to the PM upon NaCl treatment in 35S::GFP-AtCLCf roots (Fig. 7a). Hence, we used this system to test if the translocation mechanism indeed involves the exocytic trafficking pathway. After 3 h of treatment with the trafficking inhibitor brefeldin-A (BFA) alone or with BFA plus 50 mM NaCl, GFP-AtCLCf proteins aggregated with the endosomal membrane to form BFA bodies and failed to be translocated to the PM (Fig. 7a, b), which is consistent with the known effect of BFA on exocytic trafficking23,24. These results indicate that BFA prevents the translocation of AtCLCf to the PM even under salt stress. Furthermore, root elongation was reduced in seedlings treated with 5 µM BFA plus 50 mM NaCl (Fig. 7c), mimicking the phenotype of atclcf roots.
a Expression of GFP-AtCLCf in the roots of one-week-old Arabidopsis seedlings under various treatments (untreated; 50 mM NaCl; 50 µM brefeldin-A (BFA) for 3 h; 50 mM NaCl +50 µM BFA for 3 h). Scale bars, 20 µm; cor cortex, ep epidermis. b Quantification of GFP-AtCLCf expression in the PM and BFA bodies (mean ± SE, n = 15). Means with different letters within a data set are significantly different, P < 0.05. (one-way ANOVA followed by Tukey’s test). c Growth of one-week-old seedlings treated with 0 and 5 µM BFA + 50 mM NaCl. d Representative confocal images showing RFP-AtCLCf localization in untreated and 50 mM NaCl-treated WT, bex5, QL, and SN leaf protoplasts (561 nm excitation, 575–600 nm emission); scale bars, 10 µm. e, f Root lengths of one-week-old seedlings [WT, atclcf, bex5, SN, QL, and 35S::AtCLCf;atclcf (left to right) lines without (untreated) and with 50 mM NaCl]. Scale bar, 1 cm. Data are mean ± SE, n = 40. Means with different letters within a data set are significantly different, P < 0.05. (one-way ANOVA followed by Tukey’s test). g Model shows that the NaCl-induced translocation of AtCLCf from the Golgi to the PM to enhance salinity tolerance is mediated by AtRABA1b/BEX5. Data in a and d were obtained from three independent experiments, and representative images are shown here. Root tips are marked by white dots in c and e.
Recycling between the PM and endosomes is a characteristic of plant PM proteins23,24,25,26. We hypothesized that Rab GTPases, which are crucial regulators of vesicle trafficking27,28 might be involved in the translocation of AtCLCf to the PM. We focused on AtRABA1b (also called BEX5) as the potential facilitator because it was reported to be involved in the transport of cargos between the TGN and PM, as well as plant salt tolerance23,29,30. Indeed, when expressed in bex5 protoplasts, RFP-AtCLCf failed to be translocated to the PM even under NaCl treatment (Fig. 7d and Supplementary Fig. 8c), and bex5 seedlings were sensitive to BFA treatment (Fig. 7c). To further confirm that AtRABA1b is involved in the trafficking of AtCLCf, we used two AtRABA1b mutant proteins: the GTP-bound form, GFP-AtRABA1bQ72L (QL), and the GDP-bound form, GFP-AtRABA1bS27N (SN). While QL acts as a constitutively active RAB GTPase and accumulates on membranes, SN is a dominant negative form whose accumulation on membranes is significantly lower than that of WT AtRABA1b23,29. When RFP-AtCLCf was expressed in the leaf protoplasts of plants harboring QL (QL plants), significantly more AtCLCf localized to the PM than in WT protoplasts. By contrast, the localization pattern of AtCLCf in SN protoplasts was similar to that in bex5 protoplasts (Fig. 7d and Supplementary Fig. 8c). Finally, when seedlings were grown in the presence of 50 mM NaCl, the root lengths of the atclcf, bex5 and SN genotypes were significantly reduced (~40%) compared to those of untreated controls (Fig. 7e, f), whereas this treatment had less of an effect on WT, 35S::AtCLCf;atclcf and QL seedlings. A similar effect of salt was seen on the seedling root growth of SN and QL plants in an earlier study29. Collectively, these findings indicate that AtRABA1b/BEX5 facilitates the NaCl-induced translocation of AtCLCf to the PM to enhance plant salinity tolerance (Fig. 7g). This represents a unique mechanism of salt adaptation.
Discussion
Studies on the effect of salinity stress on plants have mostly focused on the involvement of Na+, while the counter-anions such as Cl− are often neglected. Hence, little is known about the molecular and cellular mechanisms of plant tolerance to Cl− ion toxicity imposed by salinity stress6. Plants respond to salinity by sequestration or efflux of ions specifically to reduce cytosolic Na+ and Cl− concentrations31,32. Several vacuolar sodium hydrogen exchangers sequester Na+ into the vacuole under salt stress33, but only Salt Overly Sensitive 1, a PM Na+/H+ antiporter, has been extensively characterized as a Na+ efflux transporter in plants34,35,36.
Accumulation of excessive Cl− reduces the growth of plants by decreasing the photosynthesis rate and leaf water potential, increasing ROS production and causing stomatal closure5,31. The effect of salt stress due to Cl− toxicity can be mitigated by means of Cl− efflux to maintain intracellular Cl− homeostasis particularly in salt-sensitive plants such as glycophytes37,38. To reduce Cl− toxicity in plants, ion channels such as members of the CLC family are predicted to be involved. The expression of GmCLC1 helped to prevent damage to cytoplasmic and organelle membranes caused by excess Cl−39. Plant CLC proteins play a major role in balancing the anion concentration under salinity. Thus, ectopic expression of some Cl− channels in targeted tissues or in whole plants have been reported to increase their salinity tolerance, but the mechanism of action is poorly understood8,40. In Arabidopsis, seven genes (AtCLCa-g) encoding CLC channel/transporter proteins have been identified. AtCLCc and AtCLCg are located in the tonoplast and are known to improve salt tolerance of plants41,42. In our study, AtCLCf enhanced the salt tolerance of both seedlings and mature plants (Figs. 1 and 2), which is in agreement with the finding that other plant Cl− transporters affect salt tolerance43,44,45.
There are no prior reports on the molecular regulation of CLC family genes thus far. Significant suppression of AtCLCf in atwrky9 mutant, and the observed interaction of AtCLCf promoter with AtWRKY9 suggest that AtWRKY9 is a key regulator of AtCLCf expression as part of the salt stress signal transduction mechanism. This conclusion is also supported by the multiple reports on the involvement of various WRKY genes in salt and drought tolerance of plants19,46,47.
It was evident from the homology model generated (Fig. 4a–c) based on the Salmonella typhimurium crystal structure48 that the conserved Cl− selectivity filter domain is also present in AtCLCf. To functionally characterize plant ion transporters, heterologous expression of Cl− and Na+ transporters in yeast mutants has been extensively used35,39,44,45. Δgef1 yeast mutant was used to understand the role of CLC transporters in regulating the ion homeostasis under salt/Cl− stress. In our study, AtCLCf was able to complement the salt sensitive phenotype of Δgef1 mutant. When some of the critical amino acid residues in two of the three selectivity filter domains were substituted [MD2: (E247W), MD3: (L463W, P466F)], their ability to complement Δgef1 was lost (Fig. 4e). Parallel to the yeast results, atclcf lines with mutation in these specific domains showed higher sensitivity towards salt and accumulated more Cl− in the protoplasts compared to AtCLCf ectopic expression lines, which indicates that these conserved domains play an important role in the protein’s function as a chloride channel. These results also imply that introducing these amino acid substitutions influences the function of AtCLCf by affecting its conformation/transport activity. Moreover, the residue that is altered in the 35S::MD2;atclcf mutant, glutamate 247 in the selectivity filter domain 2, is important for the functioning of AtCLCf, as has also been found in other organisms9,10. The observation that AtCLCf shows transport activity of H+ ions in both electrophysiological experiments and HPTS assays (Supplementary Fig. 6) suggests that it acts as a Cl−/H+ antiporter with higher specificity for Cl− transport. While some of the CLCs are reported to show NO3 transport activity, our results show that AtCLCf is not involved in NO3 transport14,15,49
Previous studies21 had indicated that AtCLCf is present within the Golgi and TGN/EE15,16,21. Our initial studies with transient assay in N. benthamiana helped to confirm this. Based on such information of subcellular localization of channels/transporters in the endosomal membranes, such as AtCLCd, AtCLCf, AtNHX5, and AtNHX6, it was interpreted that they might directly contribute to Cl− and Na+ homeostasis in the endomembrane space of plant cells under salt stress conditions50. However, our study revealed a novel mode of action of AtCLCf that involves a salt-mediated translocation of the protein from the Golgi to PM via TGN (Fig. 6). TGN is a cellular compartment where endocytic and secretory pathways intersect, and it mediates the recycling of endocytic vesicles back to the PM. This helps to explain why atclcf seedlings showed decreased Cl− efflux compared to the 35S::AtCLCf;atclcf seedlings upon salt treatment (Fig. 5). Additionally, the observed increase of AtCLCf transcripts in the roots and AtCLCf expression in the Arabidopsis root cortex/epidermis after salt treatment (Supplementary Fig. 8) further shows that AtCLCf might modulate Cl− efflux from roots under salt stress. This is supported by the observation that atclcf ISO vesicles failed to show any transport activity (Supplementary Fig. 5), along with similar data from the quantitative fluorescence assays with protoplasts (Supplementary Fig. 7). Our electrophysiological analyses in HEK293FT cells further confirm our findings that the AtCLCf is involved in Cl− efflux (Fig. 3). This efflux mechanism could prevent the accumulation of Cl− in shoots leading to increased salt tolerance observed in the ectopic expression lines (Figs. 1 and 2). The fact that BFA, a known inhibitor for protein secretion and membrane trafficking51 effectively blocked the PM translocation of AtCLCf, which partially explains why all the tested Arabidopsis genotypes showed short root phenotype upon BFA treatment. The possibility of BSA blocking other trafficking pathways such as that of auxin52 in addition to the AtCLCf translocation cannot be dismissed (Fig. 7). Also, the protoplasts from AtRABA1bQ72L plants showed increased expression of AtCLCf on the PM, further confirming that the translocation occurs through the vesicular trafficking process involving AtRABA1b/BEX5. This represents the cellular/molecular mechanism to alleviate the Cl− ion toxicity under salt stress. The finding that a chloride channel is translocated to the PM in plants has wider significance and needs to be understood in great detail because in humans, mutations in Barttin, an accessory protein required for the functioning and trafficking of a human chloride channel (CIC-K) to the PM, lead to symptoms such as renal salt loss and deafness12.
Overall, our study revealed how NaCl modulates AtCLCf activity by triggering its translocation from the Golgi apparatus to the PM to increase the Cl− efflux capacity in roots, representing an essential aspect of salt tolerance in plants. This knowledge could be used to improve salinity tolerance in crops in the future.
Methods
Plant materials and growth conditions
T-DNA insertional mutants, atclcf1 (SAIL_174_G06), atclcf2 (SALK_059540.1), and wild-type (WT) Arabidopsis thaliana, ecotype Columbia-0 were purchased from the Arabidopsis Biological Resource Centre (ABRC). The overexpression line, 35S::AtCLCf was generated in WT background, and the complementation line pAtCLCf::AtCLCf;atclcf was generated in atclcf background by us. The SN and QL lines were gifts from Dr. Tomohiro Uemura. Cold stratified Arabidopsis seeds were grown on MS medium solidified with Gelrite (0.5% w/v), with or without treatments (50 mM NaCl/KCl/NaNO3/mannitol or 5 µM BFA or 5 µM BFA + 50 mM NaCl) for 1 week and used for the following analyses: a) Root length assays from untreated and treated one-week-old seedlings were performed using Image J software53 and root length in cm was plotted as mean ± SE, n ≥ 15. b). Root fine phenotyping was carried out by treating the one-week-old seedlings with 50 mM NaCl for 6 h by submerging the seedlings with the salt solution while the seedlings were growing on the semi-solid MS plates. Following the treatment, seedlings were fixed in 1% PFA and imaged using a LEICA CTR5000 differential interference contrast (DIC) microscope equipped with a Nikon DS-Ri1 camera. Effect of NaCl treatment on meristematic zone (MZ, quiescent centre to the first elongated cortical cell), and elongation zone (EZ, first elongated cortical cell to the appearance of bulging of root hairs from the trichoblasts) were analyzed using Image J software and the length of each zone was plotted as mean ± SE, n = 10. For studies with older seedlings, one-month-old seedlings were treated with 150 mM NaCl for one week. Chlorophyll content and FW measurements were carried out as described earlier54. For survival analysis, the pots were flushed with water twice to remove the soil-bound NaCl followed by a recovery growth in NaCl-free water for one week.
In silico analysis
The NCBI database was used as a search engine for nucleotide and protein sequences. Expasy tool (https://web.expasy.org/translate/) was used to translate the CDS sequences to amino acid sequences and multAlin (http://multalin.toulouse.inra.fr/multalin/) was used to align the amino acid sequences. Phylogenetic analysis was carried out using http://www.phylogeny.fr/. Primers for qRT-PCR were designed using NCBI. Transmembrane helices of the proteins were predicted using TMHMM.
Cloning and generation of transgenic Arabidopsis lines
Mutant lines atclcf1 (SAIL_174_G06) and atclcf2 (SALK_059540.1) with T-DNA insertions were obtained from the SALK collection55. Positions of T-DNA insertion sites are shown in Supplementary Fig. 9a, b. Plants homozygous for the T-DNA insertion were selected by genotyping with primers designed using the T-DNA primer design tool (http://signal.salk.edu/tdnaprimers.2.html). Seeds were collected only from those atclcf lines that showed more than 80% suppression of AtCLCf in qRT-PCR analysis (Supplementary Fig. 9c). For generation of overexpression lines in Arabidopsis, coding sequence of AtCLCf was cloned into pGREEN binary vector. The constructs 35S::AtCLCf, 35S::GFP-AtCLCf, pAtCLCf::GUS and pAtCLCf::AtCLCf were individually electroporated into Agrobacterium tumefaciens strain GV3101:pMP90 and introduced into WT or atclcf plants by the floral dip method56. Basta-resistant T1 transgenic plants were selected after confirmation by genotyping PCR and qRT-PCR (Supplementary Fig. 9d) analyses. For chromatin immunoprecipitation (ChIP) assay, CDS of AtWRKY9, AtWRKY6 and AtWRKY33 were cloned into pGreen binary vector with hemagglutinin (HA) fusion tag. All the plasmids were sequence verified before use and the primers used in the study are listed in Supplementary Table 2.
Chromatin immunoprecipitation (ChIP) using Arabidopsis protoplasts
Mesophyll protoplasts were isolated from leaves of 3- to 4-week-old WT Arabidopsis (Col-0) plants and transfected as described earlier57 with minor modifications. For each transfection, 10 μg of purified plasmid DNA (35S::AtWRKY6, 35S::AtWRKY9 or 35S::AtWRKY33) was used. Polyethylene glycol (PEG)-CaCl2 transfection solution used was as follows: 25 % PEG, 0.4 M mannitol, and 150 mM CaCl2. The transfected protoplasts were incubated for 20 h at room temperature and fixed with formaldehyde. Protoplasts with empty plasmids were used as the negative control. ChIP was performed as described previously58, with minor changes. Anti-HA monoclonal antibody (Santa Cruz Biotechnology) bound to Protein-A agarose beads (Sigma) was used to immunoprecipitate the genomic DNA fragments. ChIP-qPCR analysis was carried out as described59 to check for enrichment of promoter fragment in the final eluted chromatin. Fold change (promoter fragment vs. no protein control) was plotted using data from 3 independent biological replicates each with three technical replicates.
Luciferase assay using Arabidopsis protoplasts
Mesophyll protoplasts were isolated from leaves of 4-week-old atwrky9 mutant seedlings, and the luciferase assay was carried out as described earlier60. 2 kb upstream sequence of AtCLCf was cloned into the pGreen II-0800-LUC vector to generate the reporter. The vector with the pAtCLCf promoter (pAtCLCf::LUC) was used as a reference control while 35S::AtWRKY9 was used as the effector. Five WRKY binding sites in the AtCLCf promoter fragment were mutated (TTGAC to TTacC) by site directed mutagenesis, and the mutant promoter was cloned into pGreen II-0800-LUC vector and used as an additional control. The luciferase assay was carried out using the Dual-Luciferase® Reporter Assay System (Promega) following the manufacturer’s instructions. The luminescence was measured using the GloMax discover (Promega). Firefly luciferase activity was normalized to Renilla luciferase activity. Data shown were taken from five independent biological replicates each with three technical replicates.
Subcellular localization of AtCLCf in N. benthamiana leaf epidermal cells
The coding sequence of GFP was fused in-frame at the N-terminal of AtCLCf in pGREEN plasmid. Empty vector, 35S::GFP-pGREEN was used as the control. These plasmids as well as the plasmids containing subcellular markers (Golgi, TGN and PM) were introduced into Agrobacterium. The subcellular marker plasmids (Golgi-rk CD3-967 and PM-rk CD3-1007) were obtained from TAIR. SYP61-RFP61 was a gift from Dr. Shen Quan Pan. Mature leaves from 3- to 4-week-old N. benthamiana plants were co-infiltrated with Agrobacterium harboring 35S::GFP-AtCLCf and the related subcellular marker constructs. N. benthamiana leaf epidermal cells were examined for GFP-AtCLCf expression along with mCherry-tagged markers using a confocal laser scanning microscope (Olympus FV3000) with 488 nm excitation, 500–525 nm emission: and 561 nm excitation, 575–600 nm emission, respectively. For salt treatment, N. benthamiana leaves were infiltrated with 100 mM NaCl and imaged after 4 and 6 h. ImageJ software was used to measure the colocalization of AtCLCf with Golgi, TGN and PM markers and plotted as Rcoloc values. The Rcoloc values were calculated using Returns Pearson’s correlation coefficient for pixels where both GFP and RFP are above their respective threshold. This generates a value for each channel. For Ch1, this value is equal to the sum of the pixel intensities with intensities above both Ch1 and Ch2 thresholds i.e. (sum of Ch1 pixel intensities in the Yellow area) ÷ (sum of Ch1 pixels intensities in the Red+Green+Yellow areas).
Subcellular localization of AtCLCf in Arabidopsis leaf protoplasts
The coding sequence of GFP or RFP were fused in-frame at the N-terminal of AtCLCf in pGREEN plasmid. Mesophyll leaf protoplasts were isolated from one-month-old plants of WT, bex5, SN and QL genotypes as described earlier57. The protoplasts were transfected with 10 µg of 35S::GFP-AtCLCf or 35S::RFP-AtCLCf using a polyethylene glycol-calcium chloride transfection solution as described previously54. After the transfection for 16 h, untreated and treated (50 mM NaCl for 3 h) protoplasts were visualized under confocal laser scanning microscope (Olympus FV3000). RFP-AtCLCf was visualized using 561 nm excitation and 600–620 nm emission. For each measurement of AtCLCf localization in PM, ~20 protoplasts from each genotype were used and the data are represented as mean ± SE.
Homology modeling of AtCLCf and site directed mutagenesis of the selectivity filter domain residues
A homology model of AtCLCf was constructed based on the crystal structure of Salmonella typhimurium CLC9, using the PyMOL software (https://pymol.org/2/). Pore lining residues present in the selectivity filter domains 1, 2 and 3 were mutated to study their role on Cl− transport function of AtCLCf. The following residues were mutated in domains 1–3 and labeled as mutated domain (MD) 1–3; MD1 (Isoleucine (I) 204 to Phenylalanine (F), Glutamine (Q) 207 to Tyrosine (Y)), MD2 (Glutamate (E) 247 to Tryptophan (W) and MD3 (Lysine (L) 463 to W, Proline (P) 466 to F). PCR-based site directed mutagenesis was carried out as described earlier62, followed by cloning into pGREEN binary vector. Using these mutant AtCLCf-pGREEN constructs, 35S::AtCLCfMD1, 35SS::AtCLCfMD2 and 35S::AtCLCfMD3 lines were generated in the atclcf background to study their response to NaCl treatment and Cl− transport functionality.
PM isolation by two-phase partitioning and formation of inside-out vesicles
The detailed methods are given in Supplementary Information. Briefly, plasma membranes (predominantly right-side-out vesicles) were purified from a microsomal fraction of 6-week-old Arabidopsis seedlings (150 g FW) by partitioning in an aqueous polymer two-phase system as described earlier63. Pelleted PM proteins were resuspended in PM washing buffer complemented with protease inhibitor and 5 mM DTT, frozen in liquid nitrogen, and stored for later use at −80 °C. This mainly consisted of the right-side-out vesicles. Using these highly purified right-side-out PM vesicles inside-out vesicles were prepared as described earlier64.
Spectrofluorimetric analysis of Cl− transport in PM vesicles
For spectrofluorimetric analysis, N-(ethoxycarbonylmethyl)-6-methoxyquinolium bromide (MQAE)-loaded ISO and RSO PM vesicles were prepared by incubation of vesicles in a hypotonic medium for 15 min at 37 °C. The hypotonic medium comprised of Hanks’ balanced salt solution (HBSS) diluted 1:1 with water and contained 5 mM MQAE. At the end of the loading period, 100 µl samples of cell suspension were diluted 15:1 in HBSS, centrifuged, resuspended in 200 µl of HBSS and then incubated for a further 15 min at 37 °C to allow recovery from the hypotonic shock. The spectrofluorimetric analysis was performed with these vesicles, stirred magnetically with a glass fly inside a standard cm path-length quartz fluorescence cuvette. A constant temperature of 20 °C was maintained by water circulating through the cuvette holder of the spectrofluorometer. Reaction was started by adding 2 ml of suspension medium (50 µl ISO/RSO MQAE-loaded PM vesicles in 5 mM phosphate buffer) was added to the cuvette. After one minute, 50 mM NaCl was added to the buffer to determine the fluorescence response. The excitation and emission wavelengths used for MQAE were 360 nm and 450 nm, respectively. The fluorescence values were normalized to 1 and plotted to show the quenching of fluorescence after addition of NaCl.
Liposome-based spectrofluorimetric analysis of Cl− transport
Codon optimization and recombinant protein expression: In order to carry out liposome-based assays, synthetic gene expression construct was prepared by cloning the codon-optimized (for yeast, Pichia pastoris), full length AtCLCf gene into yeast expression vector pPICZA (GenScript Biotech, Singapore) that was used for recombinant protein production subsequently. The original and codon-optimized sequence of AtCLCf is provided in Supplementary Fig. 10. Recombinant protein expression and purification were carried out as described in detail in Invitrogen manual (https://assets.fishersci.com/TFS-Assets/LSG/manuals/easyselect_man.pdf?_ga=2.6716082.1874667851.1688092554-921422677.1688092554). In brief, the construct was introduced into SMD1168H Pichia yeast strain using electroporation method65. A colony with the highest protein expression level was selected for further analysis. The protein expression was induced in a 500 ml BMMY culture by adding 5 % methanol and growing the culture at 28 °C for 48 h. The cells were pelleted and resuspended in solubilization buffer (20 mM Tris-HCl, pH 8.0, 20 mM K2HPO4, 300 mM NaCl, 10% glycerol) and lysed with the use of glass beads. The unlysed cells were removed by centrifugation at 4 °C, 12,000 × g for 30 min and the supernatant was applied to a HisTrap HP column (GE healthcare), and the recombinant protein was eluted by adding 500 mM imidazole. Protein expression and purity were confirmed by SDS-PAGE and western-blot analyses using anti-His (Cat # SC−8036, 1:5000) and anti-Mouse (Cat # SC-2005, 1:30000) antibodies (Santacruz) (Supplementary Fig. 11).
Preparation of AtCLCf-liposomes and spectrofluorimetric analysis: Liposomes were prepared by the film rehydration method. 25 mg of soy lipid extract was dissolved in 2 ml of chloroform. The chloroform was evaporated using a rotary evaporator in a round-bottomed flask. The dried lipid film was rehydrated with 2 ml of 5 mM sodium phosphate buffer pH 7.5, plus chloride sensitive dye MQAE. Liposomes with uniform size were obtained by passing the dye incorporated liposomes through a hand-held, extruder with polycarbonate nucleopore filters (200 nm, Whatman). The entire 2 ml solution after the extrusion process was transferred to a dialysis cassette (Thermo Scientific) and dialyzed against 5 mM sodium phosphate buffer pH 7.5 for 2 days at 4 °C to remove the excess dye. The liposome suspension was removed from the dialysis cassette and used for AtCLCf incorporation. AtCLCf-liposomes were prepared by incubating the purified AtCLCf and dye incorporated liposomes (protein-to-lipid ratio of 1:100) for 15 min at room temperature. The spectrofluorimetric analysis for Cl− transport was carried out as described for the PM vesicles above. For the HPTS assay, dried lipid was rehydrated with 2 ml of 10 mM HEPES buffer pH 7.0, 100 mM NaCl plus a pH-sensitive dye HPTS (8-hydroxy-1,3,6-pyrenetrisulfonate) The HPTS-containing liposome suspension was added to the buffer (10 mM HEPES, 100 mM NaX, pH 7.0, X = Cl−, F−, I−, NO3−) for anion selectivity assays. A pH gradient of 7–8 was applied across the vesicles by addition of NaOH, and ion transport activities were monitored by increment of HPTS fluorescence (λex = 450 nm, λem = 510 nm) over 150 s. The HPTS assays were carried out from three such independent liposome preparations.
Measurement of Cl− content in the Arabidopsis protoplasts
Leaf mesophyll protoplasts were isolated from one-month-old WT, atclcf, 35S::AtCLCf;atclcf and 35S::MD2AtCLCf;atclcf (35S::MD2) seedlings. Protoplasts treated with 50 mM NaCl for 3 h were loaded with 100 µM MQAE for 30 min and visualized under confocal microscope (Fv3000). The percentage of protoplasts showing no quenching was calculated after deducting the number of protoplasts showing no fluorescence quenching by the total number of protoplasts in a microscopic field, 10 such microscopic fields were analyzed. In addition, the extent of fluorescence quenching (equivalent to the concentration of Cl− retained inside the protoplasts) was measured from protoplasts treated with 50 mM NaCl for 3 h followed by 100 µM MQAE loading for 30 min. Assay was started with 2 ml of the suspension (500 µl of the protoplasts in 5 mM phosphate buffer). Triton X-100 (20 %) was added after 75 s and the quenching was recorded. The difference in the fluorescence value (final-initial value before adding Triton X-100) was plotted as quenched fluorescence.
RNA isolation and quantitative real-time PCR (qRT-PCR) analysis
RNA was isolated from Arabidopsis using TRIzol reagent (Thermo Fisher) following the manufacturer’s instructions54. From this, 1 µg of RNA was used and cDNA was synthesized using Maxima first strand cDNA synthesis kit for qRT-PCR (Thermo Fisher) following the manufacturer’s instructions. The qRT-PCR analyses were performed using StepOneTM Real-Time PCR machine (Applied Biosystems, Foster City, CA, USA) with the following program: 20 s at 95 °C followed by 40 cycles of 03 s at 95 °C and 30 s at 60 °C using SYBR Fast ABI Prism PCR kit from KAPA (Biosystems, Wilmington, MA, USA). The qRT-PCR data were analyzed using the StepOneTM Software (v2.1, ABI). The primers used for the qRT-PCR analysis are listed in Supplementary Table S2. Gene expression levels from three biological replicates each with three technical replicates (total n = 9) were calculated based on ∆∆CT values and represented as relative expression levels (fold change) to constitutively expressed internal control, AtUbiquitin 10.
Histochemical GUS staining
For histochemical study, transcriptional reporter line pAtCLCf::GUS was generated in WT background. To carry out GUS staining assays, cold stratified seeds were sown on MS plates. One-week-old, untreated, and salt treated (100 mM NaCl for 6 h) seedlings as well as four-week-old seedling tissues were stained in the GUS staining solution [0.1 M sodium phosphate buffer pH 7.0, 10 mM EDTA, 0.1% Triton X-100, 2 mM 5-bromo-4-chloro-3-indolyl glucuronide (X-Gluc)] for 5 min followed by overnight incubation in the dark at 37 °C without shaking. After removing staining solution several washes with 50 % ethanol were performed to remove chlorophyll until the tissues were cleared66. The images of stained whole seedlings as well as various tissue parts were recorded using an Axiocam 506 color microscope (Zeiss). GUS expression in different parts of seedlings was quantified based on the relative intensities of blue coloration using ImageJ software (https://imagej.nih.gov/ij/download.html). Data presented are mean ± SE of three biological replicates, each biological replicate consisting of at least six plants. Statistical significance was determined by unpaired Student’s t test (two-tailed).
Estimation of total Cl− ion concentration from plants
Control and salt-treated 4-week-old Arabidopsis plants were harvested and rinsed briefly with distilled water to remove surface contaminating Cl−. Pools of four plants were taken as one replicate, and three independent replicates were used to generate the mean values reported. Leaves and roots from plants were separated at collection and left to dry at 50 °C for 2 days. For determination of Cl−, the extracts were prepared by grinding 0.2 g of dry tissue with 10 ml of distilled water by incubating at 25 °C for 10 min. The homogenate was centrifuged at 3000 × g for 15 min, and the supernatant filtered through Whatman qualitative filter paper (110 mm). An aliquot of filtrate was used for Cl− estimation. The Cl− concentration was measured by anion chromatography (820 IC Separation Centre; Metrohm Ion Chromatography System, Riverview, FL, USA) and presented as mg g−1DW.
For Cl− efflux analysis, 10-day-old seedlings of WT, atclcf and 35S::AtCLCf;atclcf were treated with 100 mM NaCl in 5 ml of ½ MS liquid medium for 6 h (NaCl was not added for untreated seedlings), after which they were transferred to 5 ml of plain ½ MS liquid medium and allowed to stand for another 6 h. 2 ml of this liquid medium was taken and used to quantify the amount of Cl− effluxed into the external medium by the Arabidopsis roots. One biological replicate in each genotype consisted of 15 seedlings and five such independent biological replicates were used for the analyses.
Yeast strains and yeast complementation assay
Saccharomyces cerevisiae strain BY4741 (MATa his3Δ1 leu2Δ met15Δ ura3Δ; EUROSCARF) and its derivative: BY4741 (MATa; ura3Δ0; leu2Δ0; his3Δ1; met15Δ0; YJR040w::kanMX4) were used to carry out the complementation experiments. Three selectivity filter domain binding sites were mutated by site-directed mutagenesis [(MD1(I204F, Q207Y), MD2(E247W) and MD3(L463W, P466F)] (Supplementary Fig. 12). The coding sequence of AtCLCf and mutated AtCLCf sequences were cloned downstream of the respective promoter into the yeast multicopy vector YEp352 and the primers used for amplification are listed in Supplementary Table 2. WT transformed with empty YEp35267 plasmid was used as control. The yeast complementation assays with cells expressing AtCLCf were performed on semi-solid YNB medium containing appropriate supplements. Drop tests on YNB semi-solid medium with varying concentrations of NaCl (0.5, 1, and 1.25 M) were carried out as described earlier53.
Cell culture and transfection
Human Embryonic Kidney 293 (HEK293FT) cells were cultured in DMEM medium supplemented with 10 % fetal bovine serum and 1% penicillin and 1% streptomycin and maintained in 5% CO2 incubator at 37 °C. For transfection, cells will be seeded on the petri dishes and grown overnight. Subsequently, 1.5 µg of pIRES2-EGFP or pIRES2-EGFP-HA-AtCLCf plasmids were transfected into the cells by lipofectamine 2000. The transfected cells were incubated for 24 h in a 5% CO2 incubator at 37 °C. 24 h post transfection, the cells were split and seeded on coverslips pre-coated with poly-D lysine one day before recording.
Whole-cell patch-clamp recordings and data analysis
The cells were recorded with the internal solution (pipette solution) containing (in mM) 140 CsCl, 5 EGTA, 10 HEPES, 1 MgCl2, 0.5 Na3GTP, 4 Mg-ATP, 10 Na-phoshocreatine pH 7.4 (adjusted with KOH) and external solution containing (in mM): 10 Glucose, 125 NaCl, 25 NaHCO3, 1.25 NaH2PO4.2H20, 2.5 KCl, 1.8 CaCl2, 1 MgCl2, pH 7.4 (300–310 mOsm). To test the effect of internal and external pH on the chloride transport, in some experiments the internal or external pH was changed to pH5.5. To replace internal CsCl with CsF, CsBr, CsI, CsNO3 or CsAcetate, the internal solution (pipette solution) contains (in mM) 130 CsF, CsBr, CsI, CsNO3 or CsAcetate, 10 CsCl, 5 EGTA, 10 HEPES, 1 MgCl2, 0.5 Na3GTP, 4 Mg-ATP, 10 Na-phosphocreatine pH 7.4 (adjusted with CsOH). Whole cell recordings were performed with Multiclamp 700b amplifier (Molecular Device), low-pass filtered at 1 kHz and the series resistance was typically <10 MΩ after >50% compensation. Data was analyzed by Clampfit and Prism 10.
Western blot analysis
To confirm the expression of HA-AtCLCf (Supplementary Fig. 13), transfected HEK293FT cells were lysed with RIPA lysis buffer at 4 °C. RIPA buffer consists of 150 mM sodium chloride, 0.1% Trion X-100, 0.5% sodium deoxycholate, 0.1% SDS and 50 mM Tris, pH 8.0 supplemented with mini, EDTA-free protease inhibitor cocktail (Roche). 30 µg of protein lysate was loaded for SDS-PAGE and transfer. Rabbit (rb) anti-HA antibody (Sigma Aldrich, Cat# H6908) was used in dilution of 1:1000 and β-actin was stained with anti-β-Actin (Sigma-Aldrich, A2228) antibody in the dilution of 1:5,000 as loading control.
Statistical analysis
Data presented are the mean values ± SE or SD (as stated in the legends). Significance of difference among multiple samples was estimated by one-way ANOVA followed by Tukey’s test. Means with different letters are significantly different, P > 0.05. Binary comparisons of data were statistically analyzed by unpaired, two-tailed Student’s t test (P < 0.05 and P < 0.01).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data are available in the main text or the supplementary materials. Source data are provided with this paper.
References
Munns, R. et al. Energy costs of salt tolerance in crop plants. N. Phytol. 225, 1072–1090 (2020).
Zou, Y., Zhang, Y. & Testerink, C. Root dynamic growth strategies in response to salinity. Plant Cell Environ. 45, 695–704 (2022).
Wu, H. & Li, Z. The importance of Cl− exclusion and vacuolar Cl− sequestration: revisiting the role of Cl− transport in plant salt tolerance. Front. Plant Sci. 10, 1418 (2019).
Bazihizina, N., Colmer, T. D., Cuin, T. A., Mancuso, S. & Shabala, S. Friend or foe? Chloride patterning in halophytes. Trends Plant Sci. 24, 142–151 (2019).
Teakle, N. L. & Tyerman, S. D. Mechanisms of Cl- transport contributing to salt tolerance. Plant Cell Environ. 33, 566–589 (2010).
Geilfus, C. M. Chloride: from nutrient to toxicant. Plant Cell Physiol. 59, 877–886 (2018).
Liu, X. et al. GsCLC-c2 from wild soybean confers chloride/salt tolerance to transgenic Arabidopsis and soybean composite plants by regulating anion homeostasis. Physiol. Plant. 172, 1867–1879 (2021).
Qiu, J., Henderson, S. W., Tester, M., Roy, S. J. & Gilliham, M. SLAH1, a homologue of the slow type anion channel SLAC1, modulates shoot Cl- accumulation and salt tolerance in Arabidopsis thaliana. J. Exp. Bot. 67, 4495–4505 (2016).
Dutzler, R., Campbell, E. B., Cadene, M., Chait, B. T. & MacKinnon, R. X-ray structure of a ClC chloride channel at 3.0 Å reveals the molecular basis of anion selectivity. Nature 415, 287–294 (2002).
Jentsch, T. J. CLC chloride channels and transporters: from genes to protein structure, pathology and physiology. Crit. Rev. Biochem. Mol. Biol. 43, 3–36 (2008).
Miller, C. ClC chloride channels viewed through a transporter lens. Nature 440, 484–489 (2006).
Poroca, D. R., Pelis, R. M. & Chappe, V. M. ClC channels and transporters: Structure, physiological functions, and implications in human chloride channelopathies. Front. Pharm. 8, 151 (2017).
De Angeli, A., Zhang, J., Meyer, S. & Martinoia, E. AtALMT9 is a malate-activated vacuolar chloride channel required for stomatal opening in Arabidopsis. Nat. Commun. 4, 1804 (2013).
Barbier-Brygoo, H. et al. Anion channels/transporters in plants: from molecular bases to regulatory networks. Ann. Rev. Plant Biol. 62, 25–51 (2011).
Zifarelli, G. & Pusch, M. CLC transport proteins in plants. FEBS Lett. 584, 2122–2127 (2010).
Scholl, S., Hillmer, S., Krebs, M. & Schumacher, K. ClCd and ClCf act redundantly at the trans-Golgi network/early endosome and prevent acidification of the Golgi stack. J. Cell Sci. 134, https://doi.org/10.1242/jcs.258807 (2021).
Niu, C. F. et al. Wheat WRKY genes TaWRKY2 and TaWRKY19 regulate abiotic stress tolerance in transgenic Arabidopsis plants. Plant Cell Environ. 35, 1156–1170 (2012).
Chu, X. et al. The cotton WRKY gene GhWRKY41 positively regulates salt and drought stress tolerance in transgenic Nicotiana benthamiana. PLoS One 10, e0143022 (2015).
Liang, Q. Y. et al. Chrysanthemum WRKY gene DgWRKY5 enhances tolerance to salt stress in transgenic chrysanthemum. Sci. Rep. 7, 4799 (2017).
Krishnamurthy, P. et al. Transcriptomics analysis of salt stress tolerance in the roots of the mangrove Avicennia officinalis. Sci. Rep. 7, 10031 (2017).
Marmagne, A. et al. Two members of the Arabidopsis CLC (chloride channel) family, AtCLCe and AtCLCf, are associated with thylakoid and Golgi membranes, respectively. J. Exp. Bot. 58, 3385–3393 (2007).
Khalilova, L. A. et al. Arabidopsis thaliana mutant with T-DNA insertion in the Flot1 (At5g25250) gene promotor possesses increased resistance to NaCl. Russ. J. Plant Physiol. 67, 275–284 (2020).
Feraru, E. et al. BEX5/RabA1b regulates trans-Golgi network-to-plasma membrane protein trafficking in Arabidopsis. Plant Cell 24, 3074–3086 (2012).
Geldner, N., Friml, J., Stierhof, Y. D., Jurgens, G. & Palme, K. Auxin transport inhibitors block PIN1 cycling and vesicle trafficking. Nature 413, 425–428 (2001).
Rodriguez-Furlan, C., Minina, E. A. & Hicks, G. R. Remove, recycle, degrade: Regulating plasma membrane protein accumulation. Plant Cell 31, 2833–2854 (2019).
Schwihla, M. & Korbei, B. The beginning of the end: Initial steps in the degradation of plasma membrane proteins. Front. Plant Sci. 11, 680 (2020).
Minamino, N. & Ueda, T. RAB GTPases and their effectors in plant endosomal transport. Curr. Opin. Plant Biol. 52, 61–68 (2019).
Rutherford, S. & Moore, I. The Arabidopsis Rab GTPase family: another enigma variation. Curr. Opin. Plant Biol. 5, 518–528 (2002).
Asaoka, R. et al. Arabidopsis RABA1 GTPases are involved in transport between the trans-Golgi network and the plasma membrane, and are required for salinity stress tolerance. Plant J. 73, 240–249 (2013a).
Asaoka, R. et al. New insights into the role of Arabidopsis RABA1 GTPases in salinity stress tolerance. Plant Signal. Behav. 8, https://doi.org/10.4161/psb.25377 (2013).
Munns, R. & Tester, M. Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681 (2008).
Zhang, X. K., Zhou, Q. H., Cao, J. H. & Yu, B. J. Differential Cl−/salt tolerance and NaCl-induced alternations of tissue and cellular ion fluxes in Glycine max, Glycine soja and their hybrid seedlings. J. Agron. Crop Sci. 197, 329–339 (2011).
Jiang, X., Leidi, E. O. & Pardo, J. M. How do vacuolar NHX exchangers function in plant salt tolerance? Plant Signal. Behav. 5, 792–795 (2010).
El Mahi, H. et al. A critical role of sodium flux via the plasma membrane Na+/H+ exchanger SOS1 in the salt tolerance of rice. Plant Physiol. 180, 1046–1065 (2019).
Shi, H., Ishitani, M., Kim, C. & Zhu, J. K. The Arabidopsis thaliana salt tolerance gene SOS1 encodes a putative Na+/H+ antiporter. Proc. Natl. Acad. Sci. USA 97, 6896–6901 (2000).
Ullah, A., Dutta, D. & Fliegel, L. Expression and characterization of the SOS1 Arabidopsis salt tolerance protein. Mol. Cell. Biochem. 415, 133–143 (2016).
Wang, S. et al. Overexpression of maize chloride channel gene ZmCLC-d in Arabidopsis thaliana improved its stress resistance. Biol. Plant. 59, 55–64 (2014).
Wei, Q. J., Gu, Q. Q., Wang, N. N., Yang, C. Q. & Peng, S. A. Molecular cloning and characterization of the chloride channel gene family in trifoliate orange. Biol. Plant. 59, 645–653 (2015).
Li, W. Y. et al. Tonoplast-located GmCLC1 and GmNHX1 from soybean enhance NaCl tolerance in transgenic bright yellow (BY)-2 cells. Plant Cell Environ. 29, 1122–1137 (2006).
Cubero-Font, P. et al. Silent S-type anion channel subunit SLAH1 gates SLAH3 open for chloride root-to-shoot translocation. Curr. Biol. 26, 2213–2220 (2016).
Nguyen, C. T. et al. Characterization of the chloride channel-like, AtCLCg, involved in chloride tolerance in Arabidopsis thaliana. Plant Cell Physiol. 57, 764–775 (2016).
Wei, Q. J. et al. Overexpression of CsCLCc, a chloride channel gene from Poncirus trifoliata, enhances salt tolerance in Arabidopsis. Plant Mol. Biol. Rep. 31, 1548–1557 (2013).
Jossier, M. et al. The Arabidopsis vacuolar anion transporter, AtCLCc, is involved in the regulation of stomatal movements and contributes to salt tolerance. Plant J. 64, 563–576 (2010).
Li, B., Tester, M. & Gilliham, M. Chloride on the move. Trends Plant Sci. 22, 236–248 (2017).
Wei, P., Wang, L., Liu, A., Yu, B. & Lam, H. M. GmCLC1 confers enhanced salt tolerance through regulating chloride accumulation in soybean. Front. Plant Sci. 7, 1082 (2016).
Bai, Y., Sunarti, S., Kissoudis, C., Visser, R. G. F. & van der Linden, C. G. The role of tomato WRKY genes in plant responses to combined abiotic and biotic stresses. Front. Plant Sci. 9, 801 (2018).
He, Y. et al. Genome-wide identification and expression analysis of WRKY transcription factors under multiple stresses in Brassica napus. PLoS One 11, e0157558 (2016).
Dutzler, R. The ClC family of chloride channels and transporters. Curr. Opin. Struct. Biol. 16, 439–446 (2006).
Wei, P. et al. Identification and functional characterization of the chloride channel gene, GsCLC-c2 from wild soybean. BMC Plant Biol. 19, 121 (2019).
Nedelyaeva, O. I., Shuvalov, A. V. & Balnokin, Y. V. Chloride channels and transporters of the CLC family in plants. Russ. J. Plant Physiol. 67, 767–784 (2020).
Baral, A. et al. Salt-induced remodeling of spatially restricted clathrin-independent endocytic pathways in Arabidopsis root. Plant Cell 27, 1297–1315 (2015).
Okumura, K. et al. GNOM/FEWER ROOTS is required for the establishment of an auxin response maximum for arabidopsis lateral root initiation. Plant Cell Physiol. 54, 406–417 (2013).
Rajappa, S., Krishnamurthy, P. & Kumar, P. P. Regulation of AtKUP2 expression by bHLH and WRKY transcription factors helps to confer increased salt tolerance to Arabidopsis thaliana plants. Front. Plant Sci. 11, 1311 (2020).
Krishnamurthy, P. et al. Regulation of a cytochrome P450 gene CYP94B1 by WRKY33 transcription factor controls apoplastic barrier formation in roots to confer salt tolerance. Plant Physiol. 184, 2199–2215 (2020).
Alonso, J. M. et al. Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301, 653–657 (2003).
Clough, S. J. & Bent, A. F. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 (1998).
Yoo, S. D., Cho, Y. H. & Sheen, J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572 (2007).
Kaufmann, K. et al. Chromatin immunoprecipitation (ChIP) of plant transcription factors followed by sequencing (ChIP-SEQ) or hybridization to whole genome arrays (ChIP-CHIP). Nat. Protoc. 5, 457–472 (2010).
Krishnamurthy, P. et al. Expression of AoNHX1 increases salt tolerance of rice and Arabidopsis, and bHLH transcription factors regulate AtNHX1 and AtNHX6 in Arabidopsis. Plant Cell Rep. 38, 1299–1315 (2019).
Iwata, Y., Lee, M. H. & Koizumi, N. Analysis of a transcription factor using transient assay in Arabidopsis protoplasts. Methods Mol. Biol. 754, 107–117 (2011).
Li, X. & Pan, S. Q. Agrobacterium delivers VirE2 protein into host cells via clathrin-mediated endocytosis. Sci. Adv. 3, e1601528 (2017).
Liu, H. & Naismith, J. H. An efficient one-step site-directed deletion, insertion, single and multiple-site plasmid mutagenesis protocol. BMC Biotechnol. 8, 91 (2008).
Santoni, V. Plant plasma membrane protein extraction and solubilization for proteomic analysis. Methods Mol. Biol. 355, 93–109 (2007).
Palmgren, M. G. et al. Sealed inside-out and right-side-out plasma membrane vesicles: optimal conditions for formation and separation. Plant Physiol. 92, 871–880 (1990).
Balamurugan, V., Reddy, G. R. & Suryanarayana, V. V. S. Pichia pastoris: A notable heterologous expression system for the production of foreign proteins - Vaccines. Indian J. Biotechnol. 6, 175–186 (2007).
Ravindran, P., Verma, V., Stamm, P. & Kumar, P. P. A novel RGL2-DOF6 complex contributes to primary seed dormancy in Arabidopsis thaliana by regulating a GATA tanscription factor. Mol. Plant 10, 1307–1320 (2017).
Hill, J. E., Myers, A. M., Koerner, T. J. & Tzagoloff, A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast 2, 163–167 (1986).
Acknowledgements
The authors thank Drs. Akihiko Nakano and Tomohiro Uemura (RIKEN and Ochanomizu University, Japan) for providing plant material (seeds of GFP-RABA1bQ72L GFP-RABA1bS27N), Dr. Prakash Arumugam (SIFBI, A*STAR, Singapore) for providing the yeast strains used in this study, and Dr. Jobichen Chacko for help with homology model building. We thank Prof. Elliot Meyerowitz (Caltech) and Dr. On Sun Lau (NUS) for critical reading of our manuscript. The National University of Singapore provided partial financial support as grant number A−8000149-03-00, and PhD research scholarship to S.R.
Author information
Authors and Affiliations
Contributions
P.K., S.R. and P.P.K. conceived the research plans; all authors helped design the experiments. J.F. designed the BEX5 experiments, J.X. designed the fine phenotyping experiments. H.H. and D.Y. carried out electrophysiological experiments. S.R. and P.K. carried out all other experiments and analyzed the data. P.K. and S.R. wrote the article with contributions from all authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Frans Maathuis, Sergey Shabala and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Rajappa, S., Krishnamurthy, P., Huang, H. et al. The translocation of a chloride channel from the Golgi to the plasma membrane helps plants adapt to salt stress. Nat Commun 15, 3978 (2024). https://doi.org/10.1038/s41467-024-48234-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-48234-z
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.