Abstract
Anti-mitotic chemotherapeutic agents such as taxanes activate the spindle assembly checkpoint (SAC) to arrest anaphase onset, but taxane-exposed cells eventually undergo slippage to exit mitosis. The therapeutic efficacy of taxanes depends on whether slippage after SAC arrest culminates in continued cell survival, or in death by apoptosis. However, the mechanisms that determine these outcomes remain unclear. Here, we identify a novel role for cyclin G1 (CCNG1), an atypical cyclin. Increased CCNG1 expression accompanies paclitaxel-induced, SAC-mediated mitotic arrest, independent of p53 integrity or signaling through the SAC component, BUBR1. CCNG1 overexpression promotes cell survival after paclitaxel exposure. Conversely, CCNG1 depletion by RNA interference delays slippage and enhances paclitaxel-induced apoptosis. Consistent with these observations, CCNG1 amplification is associated with significantly shorter post-surgical survival in patients with ovarian cancer who have received adjuvant chemotherapy with taxanes and platinum compounds. Collectively, our findings implicate CCNG1 in regulating slippage and the outcome of taxane-induced mitotic arrest, with potential implications for cancer therapy.
Similar content being viewed by others
Introduction
Several widely used anti-cancer drugs, including taxanes such as paclitaxel and docetaxel, and the vinca alkaloids vinblastine or vincristine, arrest the progression of cancer cells through mitosis by interfering with the assembly or dynamics of spindle microtubules, preventing their attachment to kinetochores (Weaver and Cleveland, 2005). The failure of microtubule-kinetochore attachments prevents silencing of the spindle assembly checkpoint (SAC), the cellular machinery that blocks anaphase entry until every chromosome has received a bipolar attachment to the mitotic spindle. Until this criterion is fulfilled, the SAC machinery transmits a ‘wait-anaphase’ signal by inhibiting the anaphase-promoting complex, an E3-ubiquitin ligase that marks for destruction of several protein substrates whose elimination is necessary for anaphase onset. The ‘wait-anaphase’ signal is generated by the localization of SAC components such as BUB1, BUBR1, BUB3, MAD1 and MAD2 to kinetochores that remain unattached to spindle microtubules. It suppresses CDC20, a component of the anaphase-promoting complex that enables the specific recognition of key protein substrates (reviewed in Lew and Burke, 2003; Weaver and Cleveland, 2005; Musacchio and Salmon, 2007).
Thus, cells exposed to anti-mitotic drugs arrest in prometaphase for a prolonged period. However, they eventually exit mitosis even when inhibitory concentrations of the drug remain, a phenomenon termed adaptation or mitotic slippage (Weaver and Cleveland, 2005). There is evidence to suggest that several cellular outcomes may then follow. Cells may survive and continue cycling, sometimes with a polyploid, 4N content of DNA; stop dividing and undergo senescence; or activate pathways that lead to cell death (Rieder and Maiato, 2004). The molecules that link drug-induced mitotic arrest to these different outcomes remain largely unrecognized, despite the evidence that they critically affect the sensitivity of cancer cells to the cytotoxic or cytostatic effects of many widely used anti-cancer drugs (Shi et al., 2008). In this paper, we report the results of experiments that identify one downstream determinant of the cellular outcomes following SAC activation—the atypical cyclin G1 (CCNG1).
CCNG1 was first identified as a p53-regulated transcript induced by DNA damage (Okamoto and Beach, 1994). It contains a cyclin box near its amino-terminus, but lacks the sequence motifs, characteristic of other cyclins, which specify periodic destruction by proteolysis during the cell cycle (Tamura et al., 1993). Indeed, CCNG1 does not pair with a known cyclin-dependent kinase, and thus, its biological functions are likely to be distinct, and remain to be fully elucidated. It is known, however, that CCNG1 expression can be regulated by p53 following DNA damage, initiating a feedback loop to control p53 levels through a mechanism that involves MDM2 (Kimura and Nojima, 2002; Okamoto et al., 2002). It has been proposed that these events underpin the participation of CCNG1 in the enforcement of the p53-dependent G1-S and G2 checkpoints responsive to DNA damage.
Unexpectedly, we show here that CCNG1 accumulates in taxane-exposed cells arrested in mitosis in a manner independent of signaling through p53 or the SAC, and that it influences mitotic slippage and cell survival after taxane exposure. Interestingly, ovarian cancer patients who have received adjuvant chemotherapy with taxanes and platinum compounds exhibit poorer survival if their tumors harbor >2 copies of CCNG1 irrespective of tumor stage. Collectively, our results identify a novel CCNG1-dependent mechanism that regulates the outcome of taxane-induced mitotic arrest, and provide preliminary evidence suggesting its clinical relevance in ovarian cancer. CCNG1 may represent a novel regulator of the recently proposed but poorly characterized processes (Gascoigne and Taylor, 2009; Huang et al., 2010) that link the timing of mitotic slippage with the response of cancer cells to drug-induced mitotic arrest.
Results
Increased CCNG1 expression accompanies paclitaxel-induced SAC arrest
Asynchronous cultures of the cancer cell lines U2OS, Cal51 or HCT116 were exposed to 10 μM paclitaxel for 60 min before the drug was washed out (Sena et al., 1999; Michalakis et al., 2005). CCNG1 protein levels in cell lysates were determined by western blotting between 2 and 48 h after paclitaxel exposure (Figure 1a). These conditions of paclitaxel treatment induce an accumulation of cells in prometaphase, as marked by 4N DNA content and co-staining with the mitotic marker MPM-2 (Figure 1b). Pro-metaphase arrest was also confirmed through microscopic assessment of chromosome condensation visualized by 4′,6-diamidino-2-phenylindole staining (data not shown). MPM-2 staining increases after drug exposure in all the cell lines tested, peaking at >60% between 11 and 24 h after treatment, consistent with activation of the mitotic SAC. Although untreated cells express relatively low levels of the protein, CCNG1 protein levels increase sharply after paclitaxel exposure, exhibiting, for example, an approximately 100 × increase 16 h after exposure in the case of the HCT116 cells (Figure 1a). This increase coincides with the period of maximal mitotic arrest as determined by MPM-2 expression. CCNG1 protein levels decrease rapidly as cells undergo mitotic slippage and exit mitosis, and continue to decrease, but more slowly, over the following 48 h. Similar effects are elicited when HCT116 cells are treated with the inhibitors nocodazole or monastrol, which elicit mitotic arrest by mechanisms different from paclitaxel, suggesting that the changes in CCNG1 expression represent a general response to mitotic arrest (Supplementary Figure S1).
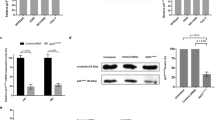
Increased CCNG1 expression accompanies paclitaxel-induced SAC-mediated mitotic arrest in a p53-independent manner. Asynchronous U2OS, Cal51 and HCT116 cells were treated with 10 μM paclitaxel for 60 min. (a) Cells were harvested over 48 h and immunoblotted with anti-CCNG1 and anti-β-actin antibodies. β-actin is used as a loading control here and in other experiments. (b) Samples of the cells described in (a) were stained with mitotic phosphoprotein monoclonal antibody, MPM-2 and propidium iodide (PI) to measure DNA content, before analysis by flow cytometry. The percentage of viable cells with 4N DNA content, or positive for MPM-2 staining, is shown. (c) Paclitaxel-induced CCNG1 expression is independent of p53. Asynchronous HCT116 cells and their isogenic p53-null counterparts (HCT116 p53−/−) were treated with paclitaxel as described before the analysis of CCNG1 and p53 expression by western blotting. (d) Samples of the cells described in (c) were fixed and stained with MPM-2 and PI as described in (b). Results shown are typical of three or more independent experiments.
Paclitaxel-induced CCNG1 expression is independent of p53
The induction of CCNG1 protein expression following cellular stresses such as DNA damage is reported to be dependent on p53 (Okamoto and Beach, 1994; Bates et al., 1996). We therefore compared CCNG1 protein levels in paclitaxel-exposed wild-type HCT116 cells with those in a p53-null, but otherwise isogenic, HCT116 derivative created by TP53 gene targeting (Bunz et al., 1998). Although p53 expression steadily increases in HCT116 cells following the treatment (Figures 1c and d), CCNG1 expression peaks at 16 h, coincident with the arrest of ∼80% of the cells in the mitotic phase of cell cycle as marked by MPM-2 positive staining, before p53 accumulation. In the p53-null HCT116 p53−/− cells, neither the kinetics nor the level of CCNG1 induction after paclitaxel exposure was changed, and cell cycle arrest in the M phase was not significantly affected. Thus, p53 is dispensable for both CCNG1 induction and mitotic arrest following paclitaxel exposure under these conditions. The p53-independence of CCNG1 induction following paclitaxel exposure was also evident when comparing the osteosarcoma cell line U2OS (p53 wild type) with its p53-deficient, non-isogenic counterpart SaOS-2 (data not shown), confirming that these findings are not confined to one particular cell type.
Paclitaxel-induced CCNG1 expression does not require signaling by the SAC
Paclitaxel alters microtubule dynamics, thereby preventing the silencing of the SAC, and thus blocking mitotic progression (Jordan and Wilson, 2004). However, several lines of evidence suggest that paclitaxel-induced CCNG1 expression does not require signaling by the SAC. First, we find that CCNG1 expression is induced by the exposure of U2OS cells to MG132 (Figure 2), a proteasome inhibitor that blocks the metaphase-anaphase transition, but without activating the SAC, by preventing the degradation of mitotic substrates required for anaphase entry (van Leuken et al., 2008). U2OS cells were exposed to 5 μM MG132 or 10 μM paclitaxel for 60 min. Ninety minutes after paclitaxel release or MG132 treatment, mitotic cells were collected by mitotic shake-off (MSO) and the level of CCNG1 expression was compared with that in untreated cells. Notably, CCNG1 expression in MG132-treated cells is similar to that in paclitaxel-treated cells, suggesting that CCNG1 induction during mitosis does not require signaling through the SAC. Similar results were obtained in Cal51 cells, suggesting that these findings are not confined to one particular cell type.
Increased CCNG1 expression during mitosis is independent of SAC activation. (a) Asynchronous U2OS cells were treated with 10 μM paclitaxel for 60 min or with 5 μM MG-132. 90 min after paclitaxel release or MG-132 treatment mitotic cells were harvested by MSO. Cell lysates were immunoblotted with anti-CCNG1 and anti-β-actin antibodies. (b) Samples of the cells described in (a) were fixed and stained with propidium iodide to measure DNA content by flow cytometric analysis. >2000 viable cells were analyzed per sample. The percentage of cells with 4N DNA content is shown. Results shown are typical of two independent experiments.
To further verify that paclitaxel-induced CCNG1 expression is independent of SAC signaling, we depleted the essential SAC component, BUBR1(Meraldi et al., 2004), from U2OS cells using siRNA, before exposure to paclitaxel as before (Figure 3a). Serial time-lapse imaging confirms that BUBR1 depletion suppresses the mitotic arrest normally triggered by paclitaxel, allowing anaphase entry and rapid exit from mitosis. Therefore, to prevent mitotic exit, BUBR1-depleted, paclitaxel-treated cells were exposed to 5 μM MG132 (Brito and Rieder, 2006), as were controls, before mitotic cells were harvested by MSO (Figure 3b). We find no detectable difference in paclitaxel-induced CCNG1 expression after BUBR1 depletion, suggesting that CCNG1 induction during mitosis is independent of signaling by the SAC (Figure 3c).
Signaling through BUBR1 is dispensable for increased CCNG1 expression during mitosis. (a) Asynchronous U2OS cells were exposed to BUBR1 or non-targeting (NT) control siRNA for 24 h before treatment with 10 μM paclitaxel for 60 min. Individual cells were visualized by bright-field microscopy every 3–5 min from entry into mitotic prophase, indicated by characteristic changes in cell morphology, until anaphase, indicated by the onset of furrowing. The median duration for this interval, and the inter-quartile range are indicated. Results shown are typical of at least two independent experiments in which 25 mitoses were followed for each experimental group. (b) Asynchronous U2OS cells were exposed to BUBR1 or NT control siRNA before paclitaxel treatment, and released into 5 μM MG132. Cells were harvested by MSO 90 min afterwards, and stained with anti-MPM-2 antibody for analysis by flow cytometry. The percentage of intact cells positive for MPM-2 staining is shown. (c) Extracts prepared from samples of the cells described in (b) were immunoblotted with anti-BUBR1, anti-CCNG1 and anti-β-actin antibodies. (d) Asynchronous Cal51 cells were treated with 10 μM paclitaxel, for 60 min, released and after 12 h, treated with 2 μM ZM 447439 (ZM). Cells were harvested 90 and 180 min after ZM addition for analysis by MPM-2 staining using flow cytometry. The percentage of viable cells positive for MPM-2 staining is shown. (e) Extracts from samples of the cells described in (d) were immunoblotted as described above. Results shown are typical of at least two independent experiments.
Notably, CCNG1 expression is decreased when paclitaxel-exposed, BUBR1-depleted cells are allowed to exit mitosis in the absence of MG132 (Figure 3c). This suggests that CCNG1 expression after paclitaxel treatment correlates with mitotic arrest rather than SAC signaling. To further confirm this, we treated Cal51 cells arrested at the metaphase-anaphase transition following paclitaxel-induced SAC activation with the Aurora kinase inhibitor ZM 447439 (ZM). ZM overrides SAC signaling, enabling mitotic exit (Ditchfield et al., 2003). Indeed, 12 h after paclitaxel treatment, Cal51 cells exposed to 2 μm ZM exit mitosis within 90 min (Figure 3d), accompanied by a decrease in CCNG1 expression to the basal levels found in unchallenged cells (Figure 3e). Similar observations were recorded in U2OS cells (data not shown). Thus collectively, our work demonstrates that paclitaxel-induced CCNG1 expression is independent of SAC signaling, but instead, accompanies the arrest during prometaphase.
CCNG1 depletion prolongs paclitaxel-induced mitotic arrest
We therefore tested whether CCNG1 regulates paclitaxel-induced mitotic arrest, using serial time-lapse imaging to precisely assess mitotic progression in CCNG1-depleted or control U2OS cells (Figures 4a and b), before and after drug exposure. Images were taken every 3 min from the start of cell rounding that starts in late prophase until the formation of a cleavage furrow in anaphase A, visualizing at least 25 individual cells per sample. The cumulative frequency of cells progressing from prophase to anaphase was then plotted against time, providing a sensitive measure of SAC enforcement. Consistent with expectation, the prophase–anaphase interval is greatly prolonged from a median of 30 min to 270 min when control cells are exposed to paclitaxel (Figure 4c). Conversely, and also as expected (Meraldi et al., 2004; Sudo et al., 2004), the depletion of BUBR1, an essential SAC component, reduces the prophase–anaphase interval after paclitaxel treatment to a median of 21 min. Interestingly, the depletion of CCNG1 using either of the two sequence-independent siRNAs significantly increased the mitotic delay induced by paclitaxel, prolonging the prophase–anaphase interval to a median of 381 min (P=0.0294) and 474 min (P<0.0001) (Figure 4d). However, CCNG1 depletion did not alter the prophase–anaphase interval in unchallenged cells, suggesting that it is dispensable for normal mitotic timing. From these results we infer that CCNG1 is not an essential component of the SAC machinery per se, but instead, may have a role in promoting slippage and mitotic exit after SAC activation.
CCNG1 depletion prolongs paclitaxel-induced mitotic arrest and increases drug-induced cell death. (a) Asynchronous U2OS and Cal51 cells exposed to CCNG1 or NT control siRNA were treated with paclitaxel, and harvested 12 h afterwards. Relative CCNG1 mRNA expression was quantitated by semi-quantitative RT–PCR. The graph shows the mean±s.e.m. from triplicate experiments. (b) Extracts from samples of the cells described in (a) were immunoblotted with anti-CCNG1 and anti-β-Actin antibodies. (c) Individual, asynchronous U2OS cells, treated as in (a), were monitored by bright-field microscopy as described in Figure 3 to determine the interval from prophase to anaphase. Cumulative frequency plots of the time taken are shown. Results shown are typical of at least two independent experiments in which 25 mitoses were followed for each experimental group. (d) The median time taken from prophase to anaphase onset after CCNG1 depletion using two distinct siRNAs is increased in a statistically significant manner (CCNG1 siRNA_1 ( ) (*P=0.0294) and CCNG1 siRNA_2 (
) (*P=0.0294) and CCNG1 siRNA_2 ( ) (***P<0.0001) when compared with NT control (
) (***P<0.0001) when compared with NT control ( ) using a two-tailed Mann–Whitney test). The median and the inter-quartile range for each group are indicated. (e) The effect of CCNG1 depletion on cell viability was assessed utilizing the CellTiter-Blue Cell Viability Assay (Promega) over a 3-day period following paclitaxel treatment. Cell viability is expressed relative to the controls. Data points represent the mean of triplicate observations with the error bars representing a single s.d. from the mean.
) using a two-tailed Mann–Whitney test). The median and the inter-quartile range for each group are indicated. (e) The effect of CCNG1 depletion on cell viability was assessed utilizing the CellTiter-Blue Cell Viability Assay (Promega) over a 3-day period following paclitaxel treatment. Cell viability is expressed relative to the controls. Data points represent the mean of triplicate observations with the error bars representing a single s.d. from the mean.
CCNG1 depletion promotes paclitaxel-induced cell death after SAC arrest
It has been proposed (Gascoigne and Taylor, 2008) that cell survival after drug-induced mitotic arrest is governed by the extent of mitotic delay, such that a prolonged delay enhances the likelihood of apoptosis. As CCNG1 depletion prolongs the prophase-anaphase interval after paclitaxel exposure, we tested its effect on paclitaxel-induced cell death. Cal51 and U2OS cells were transfected with CCNG1-specific siRNAs, and were then exposed to 10 μM paclitaxel for 60 min. Cells were harvested at 12 h after drug treatment, before the maximal induction of CCNG1 expression (Figures 4a and b), to assess CCNG1 mRNA and protein levels. The viability of the cells was then assessed by the Promega CellTiter-Blue Cell Viability Assay (Promega, Madison, WI, USA) over the following 3 days (Figure 4e). siRNA-mediated depletion under these conditions reduced CCNG1 mRNA by ∼95% (Figure 4a) and markedly lowered (but did not entirely ablate) CCNG1 protein expression (Figure 4b). CCNG1 depletion reduced the viability of both U2OS and Cal51 cells following paclitaxel exposure by 66 and 50% when compared with the controls. In all cell lines tested, reduced viability was accompanied by an increase in apoptotic caspase activity (Supplementary Figure S2). Moreover, serial time-lapse imaging suggests that CCNG1-depleted cells undergoing cell death from mitosis (or failing to complete cytokinesis and forming a single polyploid nucleus) exhibit an increase in drug-induced mitotic delay (Supplementary Figure S3). Thus, our results indicate that the prolongation of paclitaxel-induced mitotic arrest provoked by CCNG1 depletion is accompanied by an increase in drug-induced cell death.
CCNG1 over-expressing cells escape paclitaxel-induced cell death independent of p53
We therefore tested whether CCNG1 overexpression could conversely enhance cell survival after exposure to anti-mitotic drugs. To this end, we used a method we have previously established (Lee et al., 1999) to test whether cells overexpressing an EGFP-CCNG1 fusion protein have a survival advantage after paclitaxel exposure when compared with untransfected cells in the same culture. Accordingly, HCT116 cells were transfected (Figure 5a) with either EGFP-CCNG1 or EGFP, such that no more than 10–30% in each culture received the fluorophore-expressing construct, whereas the remaining cells were untransfected. Cultures were exposed to 10 μM paclitaxel for 60 min, before harvesting at different times over the following 72 h (Figure 5b), and enumeration by flow cytometry of the ratio of viable EGFP-positive to EGFP-negative cells (Figure 5c). Viable cells that express EGFP-CCNG1 are ∼2fold increased over those expressing EGFP alone within 24 h after drug treatment. This advantage was not manifest in untreated cultures confirming that EGFP-CCNG1 promotes survival only after paclitaxel exposure. Similar results were obtained in p53−/− HCT116 cells (Figure 5d), demonstrating that EGFP-CCNG1 expression promotes cell survival after SAC activation independent of p53 integrity.
CCNG1 overexpressing cells escape paclitaxel-induced apoptosis independent of p53. (a) Asynchronous HCT116 and HCT116 p53−/− cells were transiently transfected with a construct encoding an EGFP-CCNG1 fusion protein or the empty vector. Cells were treated with paclitaxel 24 h after transfection, and harvested at the indicated times for analysis by immunoblotting or flow cytometry. (b) Extracts from cells harvested at 0, 18 and 44 h were immunoblotted with anti-CCNG1 and anti-β-actin antibodies. (d) The percentage of HCT116 (c) or HCT116 p53−/− cells expressing EGFP was determined by flow cytometry. Data points represent the mean of triplicate observations with the error bars representing a single s.d. from the mean.
CCNG1 amplification is associated with significantly shorter post-surgical survival in patients with ovarian cancer who have received adjuvant chemotherapy with taxanes and platinum compounds following cytoreductive surgery
These observations prompted us to test whether CCNG1 expression might serve as a prognostic marker for patient survival in ovarian cancer, a setting in which taxanes are frequently used in adjuvant chemotherapy (Markman, 2008; Hennessy et al., 2009). We examined snap-frozen ovarian cancer tissue samples collected at debulking surgery from 100 patients, all of which were collected before treatment, for CCNG1 copy number, and the expression of CCNG1 RNA. No association was found between CCNG1 mRNA expression levels and patient survival. The lack of such an association in pre-treatment tissue samples is not unexpected, as elevated CCNG1 expression appears to be transiently induced during drug-induced mitotic arrest. Therefore, CCNG1 gene copy number was assessed by a previously reported method that is likelihood-based and explicitly takes into account changes in the total genome size, and contamination of the cancer samples with non-cancerous cells (Abkevich et al., 2010). The method is capable of detecting regions with deletions and amplifications as well as estimating actual gene copy numbers in these regions. In contrast to mRNA expression, there was a statistically significant association between CCNG1 gene copy number and patient survival. Kaplan–Meier survival plots (Figure 6) demonstrate that post-surgical survival in patients with CCNG1 copy number ⩽2 (n=66) was significantly prolonged (P=0.023) compared with that in patients with CCNG1 copy number >2 (n=34). This significance holds in multivariate Cox models after accounting for the residual size of the ovarian tumor after surgery as well as the tumor stage (P=0.009). Collectively, these data suggest that CCNG1 gene copy number is an independent marker of post-surgical survival in patients with ovarian cancer who have received adjuvant chemotherapy with taxanes and platinum compounds.
CCNG1 amplification is associated with significantly shorter post-surgical survival in patients with ovarian cancer. Kaplan–Meier survival plots show the post-surgical survival in patients with CCNG1 copy number ⩽2 (n=66) compared with that in patients with CCNG1 copy number >2 (n=34). The plots demonstrate a statistically significant difference in patient survival (P=0.023, Wilcoxon–Gehan test).
Discussion
Although it has been appreciated for some time that activation of the mitotic SAC by microtubule-disrupting agents culminates after mitotic slippage in varied biological outcomes ranging from apoptosis to survival and continued division, the mechanisms that couple checkpoint activation to these outcomes remain largely unknown. Collectively, the findings we report in this paper provide multiple lines of evidence to implicate CCNG1 as a key determinant of cell survival following SAC activation. We show that the expression of CCNG1 protein is enhanced during mitotic arrest in response to microtubule-disrupting agents (Figure 1, Supplementary Figure S1), in a manner that does not require SAC signaling, and is independent of p53 status (Figures 1,2,3). The depletion of CCNG1 by RNA interference does not itself affect normal mitotic timing in unchallenged cells, but instead, prolongs mitotic delay and reduces slippage after drug-induced SAC arrest (Figure 4). Notably, the prolongation of drug-induced mitotic arrest in CCNG1-depleted cells is accompanied by an increasing drug-induced cell death (Figure 4). Conversely, over-expression of CCNG1 promotes cell survival after paclitaxel exposure (Figure 5). Thus collectively, our findings suggest that CCNG1 acts as a determinant of the outcome of drug-induced SAC activation by regulating slippage.
A similar role for CCNG1 in determining the outcome of drug-induced SAC arrest is also evident in the diploid, non-transformed RPE1 cell line (Supplementary Figure S4), suggesting it is not restricted to cancer cells.
In contrast to prior reports, which demonstrate that p53 is required for the enhancement of CCNG1 expression after cellular stresses such as DNA damage, hypoxia or oxidative insults (Okamoto and Beach, 1994; Okamoto and Prives, 1999), we find that paclitaxel-induced CCNG1 expression is independent of p53. The p53-independence of CCNG1 induction after paclitaxel exposure is consistent with the enhancement of cell survival by CCNG1 over-expression even in p53-deficient cells; together suggesting that CCNG1 is indeed a p53-independent effector of the outcome of SAC activation.
Our findings have important implications for emerging concepts concerning the relationship between SAC slippage and cancer cell survival after anti-mitotic chemotherapy. It has recently been proposed that cell fate after a delayed mitosis is determined by the relative activity of two competing networks, respectively mediating the destruction of cyclin B1, or an increasing apoptotic signal (Gascoigne and Taylor, 2008). If cyclin B1 degradation outpaces the accumulation of the apoptotic signal, cells adapt to the checkpoint, exit into anaphase, and potentially survive. Conversely, the rapid accumulation of an apoptotic signal above the threshold required to induce apoptosis, outpacing declining cyclin B1 levels, culminates in cell death. Our findings identify CCNG1 as a novel factor that regulates this cell-fate decision, and suggest that its depletion promotes drug-induced cell death by delaying slippage and prolonging drug-induced mitotic arrest. These observations are not only consistent with the model proposed by Gascoigne and Taylor (Gascoigne and Taylor, 2008), but also provide fresh insight into the molecular mechanisms underlying it.
The clinical efficacy of anti-mitotic drugs that activate the SAC has been correlated not to their ability to induce mitotic arrest, but to their capacity to trigger drug-induced apoptosis (Shi et al., 2008). From this perspective, our work identifying CCNG1 as a determinant of the balance between cell survival/death induced by microtubule-disrupting agents has several implications for cancer therapy. CCNG1 levels may serve as a marker of the sensitivity of cancers to anti-mitotic therapy, independent of p53 mutation status. Moreover, inhibition of CCNG1 expression or activity may provide a new approach for therapeutic sensitization to these agents, again irrespective of p53 mutations. It is noteworthy in this regard that altered CCNG1 expression has been reported in cancer samples (Reimer et al., 1999; Perez et al., 2003), and that our findings suggest that CCNG1 gene amplification is an independent prognostic marker of post-surgical survival in ovarian cancer patients receiving adjuvant chemotherapy. Thus, the data we report in this paper provide a strong biologic and experimental rationale for further investigation of this connection.
Materials and methods
Cell lines, culture, and drug treatment
HCT116, U2OS and Cal51 cells were obtained either from the ATCC (American Type Culture Collection, LGC Standards, Middlesex, UK) or Cell Services (Cancer Research UK London Research Institute, London, UK). HCT116 p53−/− cells were a kind gift supplied by Dr B Vogelstein. Cells were maintained in either McCoy's 5A or DMEM media supplemented with 10% fetal bovine serum, penicillin and streptomycin as directed. Cultures were maintained at 37 °C and at 5% CO2. For paclitaxel treatment experiments, sub-confluent cells were treated with paclitaxel (Sigma, T7191, Dorset, UK) in dimethylsulfoxide. Cells were treated with 10 μM paclitaxel for 60 min before drug was washed out with media. On harvesting with trypsin, cells were collected by centrifugation in the presence of all media and phosphate buffer saline (PBS) washes used. For MG132 treatment, cells were treated with 5 μM Z-Leu-Leu-Leu-al (MG132) (Sigma, C2211) in dimethylsulfoxide. Cells were harvested, as described, after 90 min. For MSO experiments, sub-confluent cells were gently flushed with media using a 25 ml pipette at an acute angle. Mitotic cells were harvested by centrifugation of the media alone.
Cell cycle analysis and mitotic index
Both attached and floating cells were collected at the indicated time points and were centrifuged at 1000 r.p.m. for 5 min at room temperature. Cells were re-suspended in 0.5 ml PBS, fixed by adding 4.5 ml of ice-cold 70% ethanol, and incubated overnight in fixative at 4 °C. Before analysis, fixed cells were washed in PBS and incubated with propidium iodide staining solution (0.1% Triton X-100 in PBS; 0.02 mg/ml propidium iodide; 0. 2 mg/ml RnaseA) for 30 min at room temperature in the absence of light. Analysis was performed on a FACS cytometer (Becton-Dickinson, Oxford, UK). Unless otherwise stated, 10 000 events were analyzed per sample, and aggregates were excluded. To determine the mitotic fraction, cells were collected and fixed as described above. After treatment with a blocking solution (PBS containing 2% fetal bovine serum) for 1 h, cells were incubated with the MPM-2 monoclonal antibody (05-368, Upstate Technology, Billerica, MA, USA) at 1:100 in the block solution for 1 h at 37 °C, followed by an incubation with Alexa 488-conjugated goat against mouse IgG (Molecular Probes, Paisley, UK) for 1 h at RT. Cells were incubated with propidium iodide before being analyzed by FACS and the percentage of MPM-2 positive cells was determined.
Immunoblot analysis
Harvested cells were resuspended in RIPA lysis buffer (50 mM Tris (pH 8.0), 150 mM NaCl, 1% NP-40, 0.1% SDS, 0.5% sodium deoxycholate, 1 mM sodium orthovanadate, 1 mM DTT, 1 mM phenylmethylsulfonyl fluoride, and protease inhibitors (Roche, Burgess Hill, UK)). After 15 min on ice, lysates were cleared by centrifugation at 13K r.p.m. for 10 min at 4 °C. 50 μg of protein was run on a NuPAGE Novex 4–12% Bis-Tris Gel (Invitrogen, Paisley, UK). Western blot detection with antibodies described were visualized with horseradish peroxidase-coupled secondary antibody and an enhanced chemiluminescence kit (Amersham, Bucks, UK). Antibodies described include anti-CCNG1 (NCL-CYCLIN G, Novocastra, Milton Keynes, UK) at 1:200; anti-β-actin (A5441, Sigma) at 1:5000; anti-p53 (sc-126, Santa Cruz, CA, USA) at 1:1000; anti-BUBR1 (a kind gift from F. McKeon and S. Taylor) at 1:1000.
Live-cell imaging
As previously described (Daniels et al., 2004), cells were incubated in Leibovitz L15 medium (Invitrogen). Microscopy was performed using a Zeiss (Welwyn Garden City, UK) Axiovert 200 M microscope equipped with a humidified heated enclosure, and a UV filter in the excitation light path. Time-lapse images were collected every 3 to 5 min over a period of 48 h. The time taken from the start of cell rounding that starts in late prophase until the formation of a cleavage furrow in anaphase was calculated. For each condition, at least 25 individual cells were visualized.
Reverse transfection of siRNAs
Asynchronous cells were reverse transfected with 50 nM siRNA smartpool specific BUBR1 (termed BUB1B) (Dharmacon, Lafayette, CO, USA). Transfections were performed using Dharmafect 1 (Dharmacon), utilizing the standard protocols. For CCNG1 depletion, 100 nM of individual siRNAs specific for CCNG1 was reverse transfected (D-003216-05, Dharmacon—designated CCNG1_1 and S102653651, Qiagen (Crawley, UK)—designated CCNG1_2). For cyclin G1 siRNA-mediated knockdown, a second forward transfection was performed 24 h after the initial reverse transfection. AllStars Negative Control siRNA (Qiagen) were utilized as negative controls, using the same protocols as required for the target gene.
Quantitiative RT–PCR
Total RNA was extracted from cultures described by use of the Macherey-Nagel NucleoSpin RNA II Purification Kit (ABgene, Epsom, UK) with the DNase step omitted. Oligo-dT primed ss-cDNA was generated from the resulting RNA using the Cloned AMV First-Strand cDNA Synthesis Kit (Invitrogen). Q–PCR using the LightCycler 480 SYBR Green I Master mix (Roche) was performed with primers targeting the CCNG1 and PBGD mRNAs and analyzed with the use of the LightCycler 480 System (Roche). CCNG1 results were normalized to the housekeeping gene, PBGD, utilizing the ΔΔCt methodology (Schmittgen and Livak, 2008).
Cell viability
The cell viability was assessed by use of the CellTiter-Blue Cell Viability Assay (Promega). A volume of 5 μl of reagent was added directly to 100 μl of media and incubated for 90–120 min at 37 °C depending on the cell type. Data were acquired using the Fusion Universal Microplate Analyzer (Perkin Elmer, Waltham, MA, USA). Samples were excited by a Halogen source passed through a 520/10 BP filter and fluorescence was assessed for 0.1 secs/well using a 590/10 BP filter. All data had background values of reagent plus media without cells subtracted.
Expression of EGFP-CCNG1 fusion protein
A cDNA encoding human CCNG1 was isolated by RT–PCR using oligonucleotide primers that incorporated XhoI restriction sites. The resulting product after XhoI digestion was cloned into the SalI site of the pEGFP-C1 plasmid (Clontech, Saint-Germain-en-Laye, France). The empty vector control encodes enhanced green fluorescent protein alone. The pEGFP-CCNG1 plasmid and empty vector was transfected into HCT116 and HCT116 p53−/− cells using lipofectamine 2000 (Invitrogen) following standard protocols. Protein expression was determined by immunoblots as described above. Percentage of cells expressing the enhanced green fluorescent protein species was determined by flow cytometry.
RNA/DNA extraction from frozen human ovarian cancers
Sections of 10 μm thickness from 100 frozen human ovarian cancers in Tissue-Tek OCT (Qiagen, Valencia, CA, USA) were homogenized using a TissueRuptor (Qiagen) after adding QIAzol lysis reagent, followed by RNA isolation using a QIAgen miRNAeasy MiniKit per manufacturers protocol. A QIAamp DNA MiniKit (Qiagen) was used to isolate DNA per manufacturer's protocol with overnight incubation (56 °C) and RNaseA treatment.
Affymetrix 500K single-nucleotide polymorphism arrays
In 100 human ovarian cancers, 250 ng of genomic DNA was processed using GeneChip Mapping NspI or StyI Assay Kit (Affymetrix, Santa Clara, CA, USA) per manufacturer's protocol and hybridized to Affymetrix Mapping 500K NspI or StyI microarrays. After hybridization, array wash, stain and scan procedures were performed per manufacturer's protocol. Copy number analysis for CCNG1 was performed using a software package described elsewhere (Abkevich et al., 2010).
References
Abkevich V, Iliev D, Timms KM, Tran T, Skolnick M, Lanchbury JS et al. (2010). Computational method for estimating DNA copy numbers in normal samples, cancer cell lines, and solid tumors using array comparative genomic hybridization. J Biomed Biotechnol 2010: pii 386870.
Bates S, Rowan S, Vousden KH . (1996). Characterisation of human cyclin G1 and G2: DNA damage inducible genes. Oncogene 13: 1103–1109.
Brito DA, Rieder CL . (2006). Mitotic checkpoint slippage in humans occurs via cyclin B destruction in the presence of an active checkpoint. Curr Biol 16: 1194–1200.
Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown JP et al. (1998). Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science 282: 1497–1501.
Daniels MJ, Wang Y, Lee M, Venkitaraman AR . (2004). Abnormal cytokinesis in cells deficient in the breast cancer susceptibility protein BRCA2. Science 306: 876–879.
Ditchfield C, Johnson VL, Tighe A, Ellston R, Haworth C, Johnson T et al. (2003). Aurora B couples chromosome alignment with anaphase by targeting BubR1, Mad2, and Cenp-E to kinetochores. J Cell Biol 161: 267–280.
Gascoigne KE, Taylor SS . (2008). Cancer cells display profound intra- and interline variation following prolonged exposure to antimitotic drugs. Cancer Cell 14: 111–122.
Gascoigne KE, Taylor SS . (2009). How do anti-mitotic drugs kill cancer cells? J Cell Sci 122: 2579–2585.
Hennessy BT, Coleman RL, Markman M . (2009). Ovarian cancer. Lancet 374: 1371–1382.
Huang HC, Mitchison TJ, Shi J . (2010). Stochastic competition between mechanistically independent slippage and death pathways determines cell fate during mitotic arrest. PLoS One 5: e15724.
Jordan MA, Wilson L . (2004). Microtubules as a target for anticancer drugs. Nat Rev Cancer 4: 253–265.
Kimura SH, Nojima H . (2002). Cyclin G1 associates with MDM2 and regulates accumulation and degradation of p53 protein. Genes Cells 7: 869–880.
Lee H, Trainer AH, Friedman LS, Thistlethwaite FC, Evans MJ, Ponder BA et al. (1999). Mitotic checkpoint inactivation fosters transformation in cells lacking the breast cancer susceptibility gene, Brca2. Mol Cell 4: 1–10.
Lew DJ, Burke DJ . (2003). The spindle assembly and spindle position checkpoints. Annu Rev Genet 37: 251–282.
Markman M . (2008). Pharmaceutical management of ovarian cancer. Drugs 68: 771–789.
Meraldi P, Draviam VM, Sorger PK . (2004). Timing and checkpoints in the regulation of mitotic progression. Dev Cell 7: 45–60.
Michalakis J, Georgatos SD, Romanos J, Koutala H, Georgoulias V, Tsiftsis D et al. (2005). Micromolar taxol, with or without hyperthermia, induces mitotic catastrophe and cell necrosis in HeLa cells. Cancer Chemother Pharmacol 56: 615–622.
Musacchio A, Salmon ED . (2007). The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol 8: 379–393.
Okamoto K, Beach D . (1994). Cyclin G is a transcriptional target of the p53 tumor suppressor protein. Embo J 13: 4816–4822.
Okamoto K, Li H, Jensen MR, Zhang T, Taya Y, Thorgeirsson SS et al. (2002). Cyclin G recruits PP2A to dephosphorylate Mdm2. Mol Cell 9: 761–771.
Okamoto K, Prives C . (1999). A role of cyclin G in the process of apoptosis. Oncogene 18: 4606–4615.
Perez R, Wu N, Klipfel AA, Beart Jr RW . (2003). A better cell cycle target for gene therapy of colorectal cancer: cyclin G. J Gastrointest Surg 7: 884–889.
Reimer CL, Borras AM, Kurdistani SK, Garreau JR, Chung M, Aaronson SA et al. (1999). Altered regulation of cyclin G in human breast cancer and its specific localization at replication foci in response to DNA damage in p53+/+ cells. J Biol Chem 274: 11022–11029.
Rieder CL, Maiato H . (2004). Stuck in division or passing through: what happens when cells cannot satisfy the spindle assembly checkpoint. Dev Cell 7: 637–651.
Schmittgen TD, Livak KJ . (2008). Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc 3: 1101–1108.
Sena G, Onado C, Cappella P, Montalenti F, Ubezio P . (1999). Measuring the complexity of cell cycle arrest and killing of drugs: kinetics of phase-specific effects induced by taxol. Cytometry 37: 113–124.
Shi J, Orth JD, Mitchison T . (2008). Cell type variation in responses to antimitotic drugs that target microtubules and kinesin-5. Cancer Res 68: 3269–3276.
Sudo T, Nitta M, Saya H, Ueno NT . (2004). Dependence of paclitaxel sensitivity on a functional spindle assembly checkpoint. Cancer Research 64: 2502–2508.
Tamura K, Kanaoka Y, Jinno S, Nagata A, Ogiso Y, Shimizu K et al. (1993). Cyclin G: a new mammalian cyclin with homology to fission yeast Cig1. Oncogene 8: 2113–2118.
van Leuken R, Clijsters L, Wolthuis R . (2008). To cell cycle, swing the APC/C. Biochim biophys acta 1786: 49–59.
Weaver BA, Cleveland DW . (2005). Decoding the links between mitosis, cancer, and chemotherapy: the mitotic checkpoint, adaptation, and cell death. Cancer Cell 8: 7–12.
Acknowledgements
The experiments described in this paper were conceived and performed by PR and ARV, with the exception of the results in Figure 6, which were the work of BTH, JL, MSC and RCB. TF provided advice and assistance for microarray hybridization and analysis. This work was supported in ARV's laboratory by the UK Medical Research Council.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Oncogene website
Supplementary information
Rights and permissions
This work is licensed under the Creative Commons Attribution-NonCommercial-No Derivative Works 3.0 Unported License. To view a copy of this license, visit http://creativecommons.org/licenses/by-nc-nd/3.0/
About this article
Cite this article
Russell, P., Hennessy, B., Li, J. et al. Cyclin G1 regulates the outcome of taxane-induced mitotic checkpoint arrest. Oncogene 31, 2450–2460 (2012). https://doi.org/10.1038/onc.2011.431
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2011.431
Keywords
This article is cited by
-
Gastric cancer biomarker analysis in patients treated with different adjuvant chemotherapy regimens within SAMIT, a phase III randomized controlled trial
Scientific Reports (2022)
-
Mitotic checkpoint defects: en route to cancer and drug resistance
Chromosome Research (2021)
-
Atypical cyclins: the extended family portrait
Cellular and Molecular Life Sciences (2020)
-
Integrative analyses of transcriptome sequencing identify novel functional lncRNAs in esophageal squamous cell carcinoma
Oncogenesis (2017)
-
MiR-23b targets cyclin G1 and suppresses ovarian cancer tumorigenesis and progression
Journal of Experimental & Clinical Cancer Research (2016)