Abstract
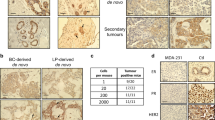
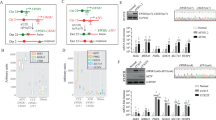
Emerging evidence suggests that cancers arise in stem/progenitor cells. Yet, the requirements for transformation of these primitive cells remains poorly understood. In this study, we have exploited the ‘mammosphere’ system that selects for primitive mammary stem/progenitor cells to explore their potential and requirements for transformation. Introduction of Simian Virus 40 Early Region and hTERT into mammosphere-derived cells led to the generation of NBLE, an immortalized mammary epithelial cell line. The NBLEs largely comprised of bi-potent progenitors with long-term self-renewal and multi-lineage differentiation potential. Clonal and karyotype analyses revealed the existence of heterogeneous population within NBLEs with varied proliferation, differentiation and sphere-forming potential. Significantly, injection of NBLEs into immunocompromised mice resulted in the generation of invasive ductal adenocarcinomas. Further, these cells harbored a sub-population of CD44+/CD24− fraction that alone had sphere- and tumor-initiating potential and resembled the breast cancer stem cell gene signature. Interestingly, prolonged in vitro culturing led to their further enrichment. The NBLE cells also showed increased expression of stemness and epithelial to mesenchymal transition markers, deregulated self-renewal pathways, activated DNA-damage response and cancer-associated chromosomal aberrations—all of which are likely to have contributed to their tumorigenic transformation. Thus, unlike previous in vitro transformation studies that used adherent, more differentiated human mammary epithelial cells our study demonstrates that the mammosphere-derived, less-differentiated cells undergo tumorigenic conversion with only two genetic elements, without requiring oncogenic Ras. Moreover, the striking phenotypic and molecular resemblance of the NBLE-generated tumors with naturally arising breast adenocarcinomas supports the notion of a primitive breast cell as the origin for this subtype of breast cancer. Finally, the NBLEs represent a heterogeneous population of cells with striking plasticity, capable of differentiation, self-renewal and tumorigenicity, thus offering a unique model system to study the molecular mechanisms involved with these processes.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 50 print issues and online access
$259.00 per year
only $5.18 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Accession codes
References
Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF . (2003). Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA 100: 3983–3988.
Ali SH, DeCaprio JA . (2001). Cellular transformation by SV40 large T antigen: interaction with host proteins. Semin Cancer Biol 11: 15–23.
Ali-Seyed M, Laycock N, Karanam S, Xiao W, Blair ET, Moreno CS . (2006). Cross-platform expression profiling demonstrates that SV40 small tumor antigen activates Notch, Hedgehog, and Wnt signaling in human cells. BMC Cancer 6: 54.
Ayyanan A, Civenni G, Ciarloni L, Morel C, Mueller N, Lefort K et al. (2006). Increased Wnt signaling triggers oncogenic conversion of human breast epithelial cells by a Notch-dependent mechanism. Proc Natl Acad Sci USA 103: 3799–3804.
Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A et al. (2008). An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet 40: 499–507.
Bhat-Nakshatri P, Appaiah H, Ballas C, Pick-Franke P, Goulet Jr R, Badve S et al. (2010). SLUG/SNAI2 and tumor necrosis factor generate breast cells with CD44+/CD24- phenotype. BMC Cancer 10: 411.
Bissell MJ, Weaver VM, Lelievre SA, Wang F, Petersen OW, Schmeichel KL . (1999). Tissue structure, nuclear organization, and gene expression in normal and malignant breast. Cancer Res 59: 1757–1763s; discussion 1763s-1764s.
Charafe-Jauffret E, Ginestier C, Birnbaum D . (2009). Breast cancer stem cells: tools and models to rely on. BMC Cancer 9: 202.
Chu PG, Weiss LM . (2002). Keratin expression in human tissues and neoplasms. Histopathology 40: 403–439.
Creighton CJ, Li X, Landis M, Dixon JM, Neumeister VM, Sjolund A et al. (2009). Residual breast cancers after conventional therapy display mesenchymal as well as tumor-initiating features. Proc Natl Acad Sci USA 106: 13820–13825.
Dey D, Saxena M, Paranjape AN, Krishnan V, Giraddi R, Kumar MV et al. (2009). Phenotypic and functional characterization of human mammary stem/progenitor cells in long term culture. PLoS One 4: e5329.
Dimri G, Band H, Band V . (2005). Mammary epithelial cell transformation: insights from cell culture and mouse models. Breast Cancer Res 7: 171–179.
Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ et al. (2003). In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev 17: 1253–1270.
Dontu G, Wicha MS . (2005). Survival of mammary stem cells in suspension culture: implications for stem cell biology and neoplasia. J Mammary Gland Biol Neoplasia 10: 75–86.
Elenbaas B, Spirio L, Koerner F, Fleming MD, Zimonjic DB, Donaher JL et al. (2001). Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes Dev 15: 50–65.
Gudjonsson T, Villadsen R, Nielsen HL, Ronnov-Jessen L, Bissell MJ, Petersen OW . (2002). Isolation, immortalization, and characterization of a human breast epithelial cell line with stem cell properties. Genes Dev 16: 693–706.
Gutierrez GM, Kong E, Sabbagh Y, Brown NE, Lee JS, Demay MB et al. (2008). Impaired bone development and increased mesenchymal progenitor cells in calvaria of RB1-/- mice. Proc Natl Acad Sci USA 105: 18402–18407.
Ince TA, Richardson AL, Bell GW, Saitoh M, Godar S, Karnoub AE et al. (2007). Transformation of different human breast epithelial cell types leads to distinct tumor phenotypes. Cancer Cell 12: 160–170.
Jerry DJ, Tao L, Yan H . (2008). Regulation of cancer stem cells by p53. Breast Cancer Res 10: 304.
Liao MJ, Zhang CC, Zhou B, Zimonjic DB, Mani SA, Kaba M et al. (2007). Enrichment of a population of mammary gland cells that form mammospheres and have in vivo repopulating activity. Cancer Res 67: 8131–8138.
Liu S, Dontu G, Wicha MS . (2005). Mammary stem cells, self-renewal pathways, and carcinogenesis. Breast Cancer Res 7: 86–95.
Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY et al. (2008). The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 133: 704–715.
Morel AP, Lievre M, Thomas C, Hinkal G, Ansieau S, Puisieux A . (2008). Generation of breast cancer stem cells through epithelial-mesenchymal transition. PLoS One 3: e2888.
Muschler J, Lochter A, Roskelley CD, Yurchenco P, Bissell MJ . (1999). Division of labor among the alpha6beta4 integrin, beta1 integrins, and an E3 laminin receptor to signal morphogenesis and beta-casein expression in mammary epithelial cells. Mol Biol Cell 10: 2817–2828.
O'Brien CA, Kreso A, Jamieson CH . (2010). Cancer stem cells and self-renewal. Clin Cancer Res 16: 3113–3120.
Pardal R, Clarke MF, Morrison SJ . (2003). Applying the principles of stem-cell biology to cancer. Nat Rev Cancer 3: 895–902.
Pardal R, Molofsky AV, He S, Morrison SJ . (2005). Stem cell self-renewal and cancer cell proliferation are regulated by common networks that balance the activation of proto-oncogenes and tumor suppressors. Cold Spring Harb Symp Quant Biol 70: 177–185.
Phesse TJ, Clarke AR . (2009). Normal stem cells in cancer prone epithelial tissues. Br J Cancer 100: 221–227.
Ponti D, Zaffaroni N, Capelli C, Daidone MG . (2006). Breast cancer stem cells: an overview. Eur J Cancer 42: 1219–1224.
Rochlitz CF, Scott GK, Dodson JM, Liu E, Dollbaum C, Smith HS et al. (1989). Incidence of activating ras oncogene mutations associated with primary and metastatic human breast cancer. Cancer Res 49: 357–360.
Rodriguez Salas N, Gonzalez Gonzalez E, Gamallo Amat C . (2010). Breast cancer stem cell hypothesis: clinical relevance (answering breast cancer clinical features). Clin Transl Oncol 12: 395–400.
Sarin KY, Cheung P, Gilison D, Lee E, Tennen RI, Wang E et al. (2005). Conditional telomerase induction causes proliferation of hair follicle stem cells. Nature 436: 1048–1052.
Shackleton M . (2010). Normal stem cells and cancer stem cells: similar and different. Semin Cancer Biol 20: 85–92.
Tan BT, Park CY, Ailles LE, Weissman IL . (2006). The cancer stem cell hypothesis: a work in progress. Lab Invest 86: 1203–1207.
Tao L, Roberts AL, Dunphy KA, Bigelow C, Yan H, Jerry DJ . (2011). Repression of mammary stem/progenitor cells by P53 is mediated by Notch and separable from apoptotic activity. Stem Cells 29: 119–127.
Uchino M, Kojima H, Wada K, Imada M, Onoda F, Satofuka H et al. (2010). Nuclear beta-catenin and CD44 upregulation characterize invasive cell populations in non-aggressive MCF-7 breast cancer cells. BMC Cancer 10: 414.
Wazer DE, Liu XL, Chu Q, Gao Q, Band V . (1995). Immortalization of distinct human mammary epithelial cell types by human papilloma virus 16 E6 or E7. Proc Natl Acad Sci USA 92: 3687–3691.
Woods C, LeFeuvre C, Stewart N, Bacchetti S . (1994). Induction of genomic instability in SV40 transformed human cells: sufficiency of the N-terminal 147 amino acids of large T antigen and role of pRB and p53. Oncogene 9: 2943–2950.
Woodward WA, Chen MS, Behbod F, Rosen JM . (2005). On mammary stem cells. J Cell Sci 118: 3585–3594.
Yang SI, Lickteig RL, Estes R, Rundell K, Walter G, Mumby MC . (1991). Control of protein phosphatase 2A by simian virus 40 small-t antigen. Mol Cell Biol 11: 1988–1995.
Zhao X, Malhotra GK, Lele SM, Lele MS, West WW, Eudy JD et al. (2010). Telomerase-immortalized human mammary stem/progenitor cells with ability to self-renew and differentiate. Proc Natl Acad Sci USA 107: 14146–14151.
Acknowledgements
We thank Dr Robert A Weinberg (Whitehead Institute) for hTERT cDNA, HMLE and HMLER cells and Dr Inder M Verma (Salk Institute) for pCSCG lentiviral vector. We acknowledge Swati Maiti for insightful discussions, and thank D Alaguraj, Drs Omana Joy, William Surin (IISc) and H Krishnamurthy (NCBS) for help with flow cytometry. We also thank Dr Paturu Kondaiah and Imran Khan (IISc), and Mohammed Aiyaz (Genotypic India Pvt. Ltd) for help with microarray experiments and data analysis. This work was largely supported by research grants from DBT to AR for equipment and consumables. We acknowledge the confocal, IRIS, FACS and animal facilities at IISc, and grants from UGC to the Department of MRDG, IISc.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no conflict of interest.
Additional information
Supplementary Information accompanies the paper on the Oncogene website
Supplementary information
Rights and permissions
About this article
Cite this article
Paranjape, A., Mandal, T., Mukherjee, G. et al. Introduction of SV40ER and hTERT into mammospheres generates breast cancer cells with stem cell properties. Oncogene 31, 1896–1909 (2012). https://doi.org/10.1038/onc.2011.378
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/onc.2011.378
Keywords
This article is cited by
-
Establishment and characterization of immortalized sweat gland myoepithelial cells
Scientific Reports (2022)
-
Roles Played by YY1 in Embryonic, Adult and Cancer Stem Cells
Stem Cell Reviews and Reports (2021)
-
Extra-telomeric functions of telomerase in the pathogenesis of Epstein-Barr virus-driven B-cell malignancies and potential therapeutic implications
Infectious Agents and Cancer (2018)
-
The viable circulating tumor cells with cancer stem cells feature, where is the way out?
Journal of Experimental & Clinical Cancer Research (2018)
-
STAT3-mediated IGF-2 secretion in the tumour microenvironment elicits innate resistance to anti-IGF-1R antibody
Nature Communications (2015)