Abstract
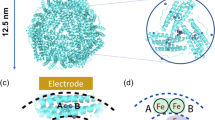
The crystal structure of Dps, a DNA-binding protein from starved E. coli that protects DMA from oxidative damage, has been solved at 1.6 Å resolution. The Dps monomer has essentially the same fold as ferritin, which forms a 24-mer with 432 symmetry, a hollow core and pores at the three-fold axes. Dps forms a dodecamer with 23 (tetrahedral) point group symmetry which also has a hollow core and pores at the three-folds. The structure suggests a novel DNA-binding motif and a mechanism for DNA protection based on the sequestration of Fe ions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout
Similar content being viewed by others
References
Farr, S.B. & Kogoma, T. Oxidative stress responses in Escherichia coli and Salmonella typhimurium. Microbiol. Rev. 55 561–585 (1991).
Almirón, M., Link, A.J., Furlong, D. & Kolter, R. A novel DNA-binding protein with regulatory and protective roles in starved Escherichia coli. Genes Dev. 6 2646–2654 (1992).
Lomovskaya, O.L., Kidwell, J.P. & Matin, A. Characterization of the σ38-dependent expression of a core Escherichia coli starvation gene, pexB. J. Bacteriol. 176 3928–3935 (1994).
Martinez, A. & Kolter, R. Protection of DMA during oxidative stress by the nonspecific DNA-binding protein Dps. J. Bacteriol. 179 5188–5194 (1997).
Altuvia, S., Almirón, M., Huisman, G., Kolter, R. & Storz, G. The dps promoter is activated by OxyR during growth and by IMF and σs during stationary phase. Mol. Microbiol. 13 265–272 (1994).
Schmid, M.B. More than just ‘histone-like’ proteins. Cell 63 451–453 (1990).
Peña, M.O. & Bullerjahn, G.S., DpsA protein of Synechococcus sp. strain PCC7942 is a DNA-binding hemoprotein. J. Biol. Chem. 270 22478–22482 (1995).
Chen, L. and Helmann, J.D. Bacillus subtilis MrgA is a Dps (PexB) homologue: evidence for metalloregulation of an oxidative stress gene. Mol. Microbiol. 18 295–300 (1995).
Bozzi, M. et al. A novel non-heme iron-binding ferritin related to the DNA-binding proteins of the Dps family in Lysteria innocua. J. Biol. Chem. 272 3259–3265 (1997).
Brentiens, R.J., Ketterer, M., Apicella, M.A. & Spinola, S.M. Fine tangled pili expressed by Haemophilus ducreyi are a novel class of pili. J. Bacterol. 178 808–816 (1996).
Pfiefer, O., Pelletier, I., Altenbuchner, J. & Van Pee, K.-H. Molecular cloning and sequencing of a non-haem bromoperoxidase gene from Streptomyces aureofaciens. J. Gen. Microbiol. 138 1123–1131 (1992).
Evans, D.J., Evans, D.G., Lampert, H.C. & Nahano, H. Identification of four new prokaryotic bacterioferritins, from Helicobacter pylori, Anabaena variabilis, Bacillus subtilis and Treponema pallidum, by analysis of gene sequences. Gene 153 123–127 (1995).
Fehniger, T.E., Radolf, J.D. & Lovett, M.A. Properties of an ordered ring structure formed by recombinant Treponema pallidum surface antigen 4D. J. Bacteriol. 165 732–739 (1986).
Sato, N. Hypothetical 20.2K protein from Anabeana variabilis. PIR accession No. JUO384 (1991).
Harrison, P.M. & Arosio, P. The ferritins: molecular properties, iron storage function and cellular regulation. Biochim. Biophys. Act. 1275 161–203 (1996).
Banyard, S.H., Stammer, D.K. & Harrison, P.M. Electron density map of apoferritin at 2.8 Å resolution. Nature 271 282–284 (1978).
Trikha, J. et al. Crystallization and structural analysis of bullfrog red-cell L-subunit ferritins. Proteins 18 107–118 (1994).
Lawson, D.M. et al. Solving the structure of human H ferritin by genetically engineering intermolecular crystal contacts. Nature 349 541–544 (1991).
Frolow, F., Kalb (Gilboa), A.J. & Yariv, J. Structure of a unique twofold symmetric haem-binding site. Nature Struct. Biol. 453–460 (1994).
Le Brun, N.E. et al. Identification of the feroxidase center of Escherichia coli bacterioferritin. Biochem. J. 312 385–392 (1995).
Nichols, A. and Honig, B. A rapid finite difference algorithm, utilizing successive over-relaxation to solve the Poisson-Boltzmann equation. J. Comput. Chem. 12 435–445 (1991).
Nichols, A., Sharp, K.A. & Honig, B. Protein folding and association: insights from interfacial and thermodynamic properties of hydrocarbons. Proteins 11 281–296 (1991).
Thompson, J.D., Higgins, D.G. & Gibson, T.J. ClustaIW: Improving the sensitivity of progressive multiple alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 4673–4680 (1994).
Rice, P.A., Yang, S.-W., Mizuuchi, K. Crystal structure of an IHF-DNA complex: a protein-induced DNA U-turn. Cell 87 1295–1306 (1996).
Shindo, H. et al. Solution structure of the DNA-binding domain of a nucleoid-associated protein, HN-S, from Escherichia coli. FEBS Lett. 360 125–131 (1995).
Tanaka, I., Appelt, K., Dijk, J., White, S.W. & Wilson, K.S. 3 Å resolution structure of a protein with histone-like properties in prokaryotes. Nature 310 376–381 (1984).
Kostrewa, D. et al. Three-dimensional structure of the E. coli DNA-binding protein FIS. Nature 349 178–180 (1992).
Yuan, H.S. et al. The molecular structure of wild-type Fis protein, relationship between mutational changes and recombinatorial enhancer function or DNA binding. Proc. Nat. Acad. Sci. USA 88 9558–9562 (1992).
Van Duyne, G.D., Standaert, R.F., Karplus, P.A., Schreiber, S.L. & Clardy, J. Atomic structures of of the human immunophilin FKBP-12 complexes with FK-506 and rapamycin. J. Mol. Biol. 229 105–124 (1993).
Ottwinowski, Z. and Minor, W. Processing of X-ray diffraction data collected in ocsillation mode. Meth. Enz. 276 307–326 (1997).
Collaborative computational project no. 4 The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. D 50 760–763 (1994).
Rould, M.A. Screening for heavy-atom derivatives and obtaining accurate isomorphous differences. Meth. Enz. 276 461–472 (1997).
Bricogne, G. Geometric sources of redundancy in intensity data and their use in phase determination. Acta Crystallogr. A30 395–495 (1974).
Filman, D.J., Wien, M.W., Cunningham, J.A., Bergelson, J.M. & Hogle, J.M. The structure determination of echovirus I. Acta Crystallogr., in the press (1998).
Jones, A.T. Interactive computer graphics: FRODO. Meth. Enz. 115 157–171 (1985).
Jones, A.T., Zou, Y-J., Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A42 140–149 (1994).
Jacobson, D.H., Hogle, J.M. & Filman, D.J. A pseudo-cell based approach to efficient crystallographic refinement of viruses. Acta Crystallogr. D 52 693–711 (1996).
Brunger, A.T. X-PLOR, Version 3.1 A system for X-ray crystallography and NMR. (Yale University Press, New Haven, Connecticut; 1992).
Holm, L. & Sander, C. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233 123–138 (1993).
deMare, F., Kurtz, D.M. Jr., Norland, P. The structure of Desulfovibrio vulgaris rubrerythrin reveals a unique combinations of rubredoxin-like FeS4 and ferritin-like diiron domains. Nature Struct. Biol. 3 539–546 (1996).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Grant, R., Filman, D., Finkel, S. et al. The crystal structure of Dps, a ferritin homolog that binds and protects DNA. Nat Struct Mol Biol 5, 294–303 (1998). https://doi.org/10.1038/nsb0498-294
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nsb0498-294
This article is cited by
-
Determination of DNA architecture of bacteria under various types of stress, methodological approaches, problems, and solutions
Biophysical Reviews (2023)
-
Linocin M18 protein from the insect pathogenic bacterium Brevibacillus laterosporus isolates
Applied Microbiology and Biotechnology (2023)
-
Protein interface redesign facilitates the transformation of nanocage building blocks to 1D and 2D nanomaterials
Nature Communications (2021)
-
Dps protein is related to resistance of Mycobacterium abscessus subsp. massiliense against stressful conditions
Applied Microbiology and Biotechnology (2020)
-
Disclosing the Molecular Mechanism of Iron Incorporation in Listeria innocua Dps by EPR Spectroscopy
Applied Magnetic Resonance (2020)