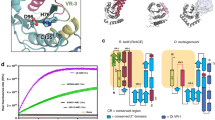
Abstract
Bacterial pathogens have evolved effector proteins with ubiquitin E3 ligase activities through structural mimicking. Here we report the crystal structure of the Shigella flexneri type III effector IpaH3, a member of the leucine-rich repeat (LRR)-containing bacterial E3 family. The LRR domain is structurally similar to Yersinia pestis YopM and potentially binds to substrates. The structure of the C-terminal E3 domain differs from the typical RING- and HECT-type E3s. IpaH3 synthesizes a Lys48-linked ubiquitin chain, and the reaction requires noncovalent binding between ubiquitin and a specific E2, UbcH5. Free ubiquitin serves as an acceptor for IpaH3-catalyzed ubiquitin transfer. Cys363 within a conserved CXD motif acts as a nucleophile to catalyze ubiquitin transfer through a transthiolation reaction. The D365N mutant is devoid of E3 activities but turns into a potent ubiquitin-E2 thioesterase. Our analysis establishes a structurally and mechanistically distinct class of ubiquitin ligases found exclusively in pathogenic or symbiotic bacteria.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
Accession codes
References
Kerscher, O., Felberbaum, R. & Hochstrasser, M. Modification of proteins by ubiquitin and ubiquitin-like proteins. Annu. Rev. Cell Dev. Biol. 22, 159–180 (2006).
Kee, Y. & Huibregtse, J.M. Regulation of catalytic activities of HECT ubiquitin ligases. Biochem. Biophys. Res. Commun. 354, 329–333 (2007).
Rytkonen, A. & Holden, D.W. Bacterial interference of ubiquitination and deubiquitination. Cell Host Microbe 1, 13–22 (2007).
Munro, P., Flatau, G. & Lemichez, E. Bacteria and the ubiquitin pathway. Curr. Opin. Microbiol. 10, 39–46 (2007).
Angot, A., Vergunst, A., Genin, S. & Peeters, N. Exploitation of eukaryotic ubiquitin signaling pathways by effectors translocated by bacterial type III and type IV secretion systems. PLoS Pathog. 3, e3 (2007).
Janjusevic, R., Abramovitch, R.B., Martin, G.B. & Stebbins, C.E. A bacterial inhibitor of host programmed cell death defenses is an E3 ubiquitin ligase. Science 311, 222–226 (2006).
Rosebrock, T.R. et al. A bacterial E3 ubiquitin ligase targets a host protein kinase to disrupt plant immunity. Nature 448, 370–374 (2007).
Zhang, Y., Higashide, W.M., McCormick, B.A., Chen, J. & Zhou, D. The inflammation-associated Salmonella SopA is a HECT-like E3 ubiquitin ligase. Mol. Microbiol. 62, 786–793 (2006).
Kubori, T., Hyakutake, A. & Nagai, H. Legionella translocates an E3 ubiquitin ligase that has multiple U-boxes with distinct functions. Mol. Microbiol. 67, 1307–1319 (2008).
Diao, J., Zhang, Y., Huibregtse, J.M., Zhou, D. & Chen, J. Crystal structure of SopA, a Salmonella effector protein mimicking a eukaryotic ubiquitin ligase. Nat. Struct. Mol. Biol. 15, 65–70 (2008).
Rohde, J.R., Breitkreutz, A., Chenal, A., Sansonetti, P.J. & Parsot, C. Type III secretion effectors of the IpaH family are E3 ubiquitin ligases. Cell Host Microbe 1, 77–83 (2007).
Haraga, A. & Miller, S.I. A Salmonella type III secretion effector interacts with the mammalian serine/threonine protein kinase PKN1. Cell. Microbiol. 8, 837–846 (2006).
Ashida, H., Toyotome, T., Nagai, T. & Sasakawa, C. Shigella chromosomal IpaH proteins are secreted via the type III secretion system and act as effectors. Mol. Microbiol. 63, 680–693 (2007).
Brzovic, P.S., Lissounov, A., Christensen, D.E., Hoyt, D.W. & Klevit, R.E.A. UbcH5/ubiquitin noncovalent complex is required for processive BRCA1-directed ubiquitination. Mol. Cell 21, 873–880 (2006).
Hochstrasser, M. Lingering mysteries of ubiquitin-chain assembly. Cell 124, 27–34 (2006).
Kobe, B. & Kajava, A.V. The leucine-rich repeat as a protein recognition motif. Curr. Opin. Struct. Biol. 11, 725–732 (2001).
Evdokimov, A.G., Anderson, D.E., Routzahn, K.M. & Waugh, D.S. Unusual molecular architecture of the Yersinia pestis cytotoxin YopM: a leucine-rich repeat protein with the shortest repeating unit. J. Mol. Biol. 312, 807–821 (2001).
Holm, L. & Sander, C. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 233, 123–138 (1993).
Huang, L. et al. Structure of an E6AP-UbcH7 complex: insights into ubiquitination by the E2–E3 enzyme cascade. Science 286, 1321–1326 (1999).
Ogunjimi, A.A. et al. Regulation of Smurf2 ubiquitin ligase activity by anchoring the E2 to the HECT domain. Mol. Cell 19, 297–308 (2005).
Verdecia, M.A. et al. Conformational flexibility underlies ubiquitin ligation mediated by the WWP1 HECT domain E3 ligase. Mol. Cell 11, 249–259 (2003).
Zheng, N., Wang, P., Jeffrey, P.D. & Pavletich, N.P. Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell 102, 533–539 (2000).
Tu, D., Li, W., Ye, Y. & Brunger, A.T. Structure and function of the yeast U-box-containing ubiquitin ligase Ufd2p. Proc. Natl. Acad. Sci. USA 104, 15599–15606 (2007).
Zhang, M. et al. Chaperoned ubiquitylation–crystal structures of the CHIP U box E3 ubiquitin ligase and a CHIP-Ubc13-Uev1a complex. Mol. Cell 20, 525–538 (2005).
McDonald, C., Vacratsis, P.O., Bliska, J.B. & Dixon, J.E. The Yersinia virulence factor YopM forms a novel protein complex with two cellular kinases. J. Biol. Chem. 278, 18514–18523 (2003).
Collaborative Computational Project, Number 4. The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D Biol. Crystallogr. 50, 760–763 (1994).
Terwilliger, T.C. & Berendzen, J. Automated MAD and MIR structure solution. Acta Crystallogr. D Biol. Crystallogr. 55, 849–861 (1999).
Terwilliger, T.C. Maximum-likelihood density modification. Acta Crystallogr. D Biol. Crystallogr. 56, 965–972 (2000).
Terwilliger, T.C. Automated main-chain model building by template matching and iterative fragment extension. Acta Crystallogr. D Biol. Crystallogr. 59, 38–44 (2003).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 (2004).
Jones, T.A., Zou, J.Y., Cowan, S.W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991).
Li, W., Tu, D., Brunger, A.T. & Ye, Y. A ubiquitin ligase transfers preformed polyubiquitin chains from a conjugating enzyme to a substrate. Nature 446, 333–337 (2007).
Acknowledgements
We thank Y. Yamada and his staff at the KEK Synchrotron Facility (Tsukuba, Japan) and Y. Dong at the Beijing Synchrotron Radiation Facility (BSRF) for assistances with data collection. We thank C. Long and S. Chen for in-house MS analysis and M.W. Silby (Tufts University) for providing the genomic DNA from P. fluorescens PfO-1. We also thank members of the Shao laboratory for helpful discussions and technical assistance. This work was supported by Chinese Ministry of Science and Technology Grant 2005AA210950 and National Basic Research Plan of China grant (2006CB806502) to F.S.
Author information
Authors and Affiliations
Contributions
Y.Z. performed the crystallography work with assistance from L.H. as well as some of the biochemical assays. H.L. performed most of the biochemical experiments. J.W., Y.Z., Z.P. and L.L. contributed to the generation of reagents. Y.Z., H.L. and F.S. designed the study, interpretated the results and prepared the manuscript.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–6 (PDF 1485 kb)
Rights and permissions
About this article
Cite this article
Zhu, Y., Li, H., Hu, L. et al. Structure of a Shigella effector reveals a new class of ubiquitin ligases. Nat Struct Mol Biol 15, 1302–1308 (2008). https://doi.org/10.1038/nsmb.1517
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb.1517