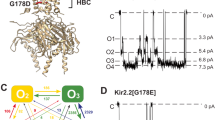
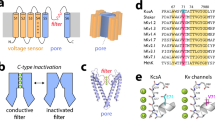
Abstract
K+ channels undergo a time-dependent slow inactivation process that plays a key role in modulating cellular excitability. Here we show that in the prokaryotic proton-gated K+ channel KcsA, the number and strength of hydrogen bonds between residues in the selectivity filter and its adjacent pore helix determine the rate and extent of C-type inactivation. Upon channel activation, the interaction between residues at positions Glu71 and Asp80 promotes filter constriction parallel to the permeation pathway, which affects K+-binding sites and presumably abrogates ion conduction. Coupling between these two positions results in a quantitative correlation between their interaction strength and the stability of the inactivated state. Engineering of these interactions in the eukaryotic voltage-dependent K+ channel Kv1.2 suggests that a similar mechanistic principle applies to other K+ channels. These observations provide a plausible physical framework for understanding C-type inactivation in K+ channels.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout








Similar content being viewed by others
References
Hoshi, T., Zagotta, W.N. & Aldrich, R.W. Two types of inactivation in Shaker K+ channels: effects of alterations in the carboxy-terminal region. Neuron 7, 547–556 (1991).
Yellen, G. The moving parts of voltage-gated ion channels. Q. Rev. Biophys. 31, 239–295 (1998).
Yellen, G., Sodickson, D., Chen, T.Y. & Jurman, M.E. An engineered cysteine in the external mouth of a K+ channel allows inactivation to be modulated by metal binding. Biophys. J. 66, 1068–1075 (1994).
Liu, Y., Jurman, M.E. & Yellen, G. Dynamic rearrangement of the outer mouth of a K+ channel during gating. Neuron 16, 859–867 (1996).
Kiss, L., LoTurco, J. & Korn, S.J. Contribution of the selectivity filter to inactivation in potassium channels. Biophys. J. 76, 253–263 (1999).
Choi, K.L., Aldrich, R.W. & Yellen, G. Tetraethylammonium blockade distinguishes two inactivation mechanisms in voltage-activated K+ channels. Proc. Natl. Acad. Sci. USA 88, 5092–5095 (1991).
Lopez-Barneo, J., Hoshi, T., Heinemann, S.H. & Aldrich, R.W. Effects of external cations and mutations in the pore region on C-type inactivation of Shaker potassium channels. Receptors Channels 1, 61–71 (1993).
Demo, S.D. & Yellen, G. Ion effects on gating of the Ca2+-activated K+ channel correlate with occupancy of the pore. Biophys. J. 61, 639–648 (1992).
Kurata, H.T. & Fedida, D. A structural interpretation of voltage-gated potassium channel inactivation. Prog. Biophys. Mol. Biol. 92, 185–208 (2006).
Seebohm, G., Sanguinetti, M.C. & Pusch, M. Tight coupling of rubidium conductance and inactivation in human KCNQ1 potassium channels. J. Physiol. (Lond.) 552, 369–378 (2003).
Chapman, M.L., Blanke, M.L., Krovetz, H.S. & Vandongen, A.M. Allosteric effects of external K+ ions mediated by the aspartate of the GYGD signature sequence in the Kv2.1 K+ channel. Pflugers Arch. 451, 776–792 (2006).
Ficker, E., Jarolimek, W., Kiehn, J., Baumann, A. & Brown, A.M. Molecular determinants of dofetilide block of HERG K+ channels. Circ. Res. 82, 386–395 (1998).
Yifrach, O. & MacKinnon, R. Energetics of pore opening in a voltage-gated K+ channel. Cell 111, 231–239 (2002).
Lu, T. et al. Probing ion permeation and gating in a K+ channel with backbone mutations in the selectivity filter. Nat. Neurosci. 4, 239–246 (2001).
Alagem, N., Yesylevskyy, S. & Reuveny, E. The pore helix is involved in stabilizing the open state of inwardly rectifying K+ channels. Biophys. J. 85, 300–312 (2003).
Gao, L., Mi, X., Paajanen, V., Wang, K. & Fan, Z. Activation-coupled inactivation in the bacterial potassium channel KcsA. Proc. Natl. Acad. Sci. USA 102, 17630–17635 (2005).
Cordero-Morales, J.F. et al. Molecular determinants of gating at the potassium-channel selectivity filter. Nat. Struct. Mol. Biol. 13, 311–318 (2006).
Zhou, Y., Morais-Cabral, J.H., Kaufman, A. & MacKinnon, R. Chemistry of ion coordination and hydration revealed by a K+ channel-Fab complex at 2.0 Å resolution. Nature 414, 43–48 (2001).
Cordero-Morales, J.F., Cuello, L.G. & Perozo, E. Voltage-dependent gating at the KcsA selectivity filter. Nat. Struct. Mol. Biol. 13, 319–322 (2006).
Kuo, A. et al. Crystal structure of the potassium channel KirBac1.1 in the closed state. Science 300, 1922–1926 (2003).
Yang, J., Yu, M., Jan, Y.N. & Jan, L.Y. Stabilization of ion selectivity filter by pore loop ion pairs in an inwardly rectifying potassium channel. Proc. Natl. Acad. Sci. USA 94, 1568–1572 (1997).
Perozo, E., Cortes, D.M. & Cuello, L.G. Three-dimensional architecture and gating mechanism of a K+ channel studied by EPR spectroscopy. Nat. Struct. Biol. 5, 459–469 (1998).
Perozo, E., Cortes, D.M. & Cuello, L.G. Structural rearrangements underlying K+-channel activation gating. Science 285, 73–78 (1999).
Baukrowitz, T. & Yellen, G. Modulation of K+ current by frequency and external [K+]: a tale of two inactivation mechanisms. Neuron 15, 951–960 (1995).
Panyi, G. & Deutsch, C. Cross talk between activation and slow inactivation gates of Shaker potassium channels. J. Gen. Physiol. 128, 547–559 (2006).
Smith, P.L., Baukrowitz, T. & Yellen, G. The inward rectification mechanism of the HERG cardiac potassium channel. Nature 379, 833–836 (1996).
Torrie, G.M. & Valleau, J.P. Nonphysical sampling distributions in Monte Carlo free-energy estimation: umbrella sampling. J. Comp. Phys. 23, 187–199 (1977).
Lenaeus, M.J., Vamvouka, M., Focia, P.J. & Gross, A. Structural basis of TEA blockade in a model potassium channel. Nat. Struct. Mol. Biol. 12, 454–459 (2005).
Berneche, S. & Roux, B. A gate in the selectivity filter of potassium channels. Structure 13, 591–600 (2005).
Stuhmer, W. et al. Molecular basis of functional diversity of voltage-gated potassium channels in mammalian brain. EMBO J. 8, 3235–3244 (1989).
Bean, B.P. The action potential in mammalian central neurons. Nat. Rev. Neurosci. 8, 451–465 (2007).
Spector, P.S., Curran, M.E., Zou, A., Keating, M.T. & Sanguinetti, M.C. Fast inactivation causes rectification of the IKr channel. J. Gen. Physiol. 107, 611–619 (1996).
Sun, Z.P., Akabas, M.H., Goulding, E.H., Karlin, A. & Siegelbaum, S.A. Exposure of residues in the cyclic nucleotide-gated channel pore: P region structure and function in gating. Neuron 16, 141–149 (1996).
Bruening-Wright, A., Schumacher, M.A., Adelman, J.P. & Maylie, J. Localization of the activation gate for small conductance Ca2+-activated K+ channels. J. Neurosci. 22, 6499–6506 (2002).
Claydon, T.W., Makary, S.Y., Dibb, K.M. & Boyett, M.R. The selectivity filter may act as the agonist-activated gate in the G protein-activated Kir3.1/Kir3.4 K+ channel. J. Biol. Chem. 278, 50654–50663 (2003).
Blunck, R., Cordero-Morales, J.F., Cuello, L.G., Perozo, E. & Bezanilla, F. Detection of the opening of the bundle crossing in KcsA with fluorescence lifetime spectroscopy reveals the existence of two gates for ion conduction. J. Gen. Physiol. 128, 569–581 (2006).
Swenson, R.P., Jr & Armstrong, C.M. K+ channels close more slowly in the presence of external K+ and Rb+. Nature 291, 427–429 (1981).
Spruce, A.E., Standen, N.B. & Stanfield, P.R. Rubidium ions and the gating of delayed rectifier potassium channels of frog skeletal muscle. J. Physiol. (Lond.) 411, 597–610 (1989).
Chapman, M.L., VanDongen, H.M. & VanDongen, A.M. Activation-dependent subconductance levels in the drk1 K channel suggest a subunit basis for ion permeation and gating. Biophys. J. 72, 708–719 (1997).
Zheng, J. & Sigworth, F.J. Intermediate conductances during deactivation of heteromultimeric Shaker potassium channels. J. Gen. Physiol. 112, 457–474 (1998).
Proks, P., Capener, C.E., Jones, P. & Ashcroft, F.M. Mutations within the P-loop of Kir6.2 modulate the intraburst kinetics of the ATP-sensitive potassium channel. J. Gen. Physiol. 118, 341–353 (2001).
Lockless, S.W., Zhou, M. & MacKinnon, R. Structural and thermodynamic properties of selective ion binding in a K+ channel. PLoS Biol. 5, e121 (2007).
Claydon, T.W. et al. A direct demonstration of closed-state inactivation of K+ channels at low pH. J. Gen. Physiol. 129, 437–455 (2007).
Cortes, D.M. & Perozo, E. Structural dynamics of the Streptomyces lividans K+ channel (SKC1): oligomeric stoichiometry and stability. Biochemistry 36, 10343–10352 (1997).
Cuello, L.G., Romero, J.G., Cortes, D.M. & Perozo, E. pH-dependent gating in the Streptomyces lividans K+ channel. Biochemistry 37, 3229–3236 (1998).
Cortes, D.M., Cuello, L.G. & Perozo, E. Molecular architecture of full-length KcsA: role of cytoplasmic domains in ion permeation and activation gating. J. Gen. Physiol. 117, 165–180 (2001).
Otwinowski, Z. & Minor, W. Macromolecular crystallography, part A. Methods Enzymol. 276, 307–326 (1997).
Jones, T.A., Zou, J.-Y., Cowan, S.W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991).
Brunger, A.T. et al. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 (1998).
Brooks, B.R. et al. CHARMM: a program for macromolecular energy, minimization, and dynamics calculations. J. Comput. Chem. 4, 187–217 (1983).
Berneche, S. & Roux, B. Molecular dynamics of the KcsA K+ channel in a bilayer membrane. Biophys. J. 78, 2900–2917 (2000).
Kumar, S., Bouzida, D., Swendsen, R.H., Kollman, P.A. & Rosenberg, J.M. The weighted histogram analysis method for free-energy calculations on biomolecules. I. The methods. J. Comput. Chem. 13, 1011–1021 (1992).
Acknowledgements
We thank F. Bezanilla, S. Chakrapani, L. Cuello, M. Sotomayor and H. Raghuraman for critical reading and discussion of the manuscript; M. Wiener and M. Purdy for crystallographic data collection (for E71T); J. Faraldo-Gomez and A. Lau for critical comments on the PMF calculations; S. Goldstein (University of Chicago) for providing access to the two electrode voltage clamp system; the staff of the GM/CA-23-ID beamline at the Advanced Photon Source for their invaluable assistance in data collection; R. MacKinnon (Rockefeller University) for providing the KcsA antibody hybridoma cell line; and the National Center for Supercomputing Applications and Jazz computing cluster at Argonne National Laboratory for computer time. This work was supported by US National Institutes of Health grants to E.P. and B.R.
Author information
Authors and Affiliations
Contributions
J.F.C.-M. carried out the channel mutagenesis, biochemistry, EPR studies, crystallization and electrophysiology with KcsA channels, and J.F.C.-M. and A.L. carried these out on Kv1.2 channels. V.J. carried out crystal data collection, structure solving and computational analyses. V.V. participated in channel mutagenesis, biochemistry and EPR. D.M.C. made all the Fab preparations. B.R., working with V.J., participated in computational design and PMF calculations. E.P. directed experimental design and data analyses, and wrote the manuscript with J.F.C.-M., V.J. and B.R.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figure 1–6, Supplementary Table 1 (PDF 4408 kb)
Supplementary Video 1
E71H movie. These movies were made with VMD. VMD is developed with NIH support by the Theoretical and Computational Biophysics group at the Beckman Institute, University of Illinois at Urbana-Champaign. (http://www.ks.uiuc.edu/)7. All the molecular graphic figures in this work were made using Pymol (DeLano, W.L. The PyMOL Molecular Graphics System (2002) http://pymol.sourceforge.net/). (MPG 3664 kb)
Supplementary Video 2
E71A movie. These movies were made with VMD. VMD is developed with NIH support by the Theoretical and Computational Biophysics group at the Beckman Institute, University of Illinois at Urbana-Champaign. (http://www.ks.uiuc.edu/)7. All the molecular graphic figures in this work were made using Pymol (DeLano, W.L. The PyMOL Molecular Graphics System (2002) http://pymol.sourceforge.net/). (MPG 3445 kb)
Rights and permissions
About this article
Cite this article
Cordero-Morales, J., Jogini, V., Lewis, A. et al. Molecular driving forces determining potassium channel slow inactivation. Nat Struct Mol Biol 14, 1062–1069 (2007). https://doi.org/10.1038/nsmb1309
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb1309
This article is cited by
-
Shifts in the selectivity filter dynamics cause modal gating in K+ channels
Nature Communications (2019)
-
Differential effect of Androctonus australis hector venom components on macrophage KV channels: electrophysiological characterization
European Biophysics Journal (2019)
-
Expression and Purification of the Pain Receptor TRPV1 for Spectroscopic Analysis
Scientific Reports (2017)
-
Discovery and characterisation of a novel toxin from Dendroaspis angusticeps, named Tx7335, that activates the potassium channel KcsA
Scientific Reports (2016)
-
Arrhythmogenic effects of mutated L-type Ca2+-channels on an optogenetically paced muscular pump in Caenorhabditis elegans
Scientific Reports (2015)