Abstract
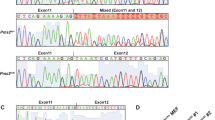
Peutz-Jeghers syndrome (PJS) is an autosomal dominant disorder associated with gastrointestinal polyposis and an increased cancer risk. PJS is caused by germline mutations in the tumor suppressor gene LKB1. One such mutation, IVS2+1A>G, alters the second intron 5′ splice site, which has sequence features of a U12-type AT-AC intron. We report that in patients, LKB1 RNA splicing occurs from the mutated 5′ splice site to several cryptic, noncanonical 3′ splice sites immediately adjacent to the normal 3′ splice site. In vitro splicing analysis demonstrates that this aberrant splicing is mediated by the U12-dependent spliceosome. The results indicate that the minor spliceosome can use a variety of 3′ splice site sequences to pair to a given 5′ splice site, albeit with tight constraints for maintaining the 3′ splice site position. The unusual splicing defect associated with this PJS-causing mutation uncovers differences in splice-site recognition between the major and minor pre-mRNA splicing pathways.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Yoo, L.I., Chung, D.C. & Yuan, J. LKB1—a master tumour suppressor of the small intestine and beyond. Nat. Rev. Cancer 2, 529–535 (2002).
Lim W. et al. Further observations on LKB1/STK11 status and cancer risk in Peutz-Jeghers syndrome. Br. J. Cancer 89, 308–313 (2003).
Hemminki, A. et al. A serine/threonine kinase gene defective in Peutz-Jeghers syndrome. Nature 391, 184–187 (1998).
Jenne, D.E. et al. Peutz-Jeghers syndrome is caused by mutations in a novel serine threonine kinase. Nat. Genet. 18, 38–43 (1998).
Bardeesy, N. et al. Loss of the Lkb1 tumour suppressor provokes intestinal polyposis but resistance to transformation. Nature 419, 162–167 (2002).
Shaw, R.J. et al. The tumor suppressor LKB1 kinase directly activates AMP-activated kinase and regulates apoptosis in response to energy stress. Proc. Natl. Acad. Sci. USA 101, 3329–3335 (2004).
Lizcano, J.M. et al. LKB1 is a master kinase that activates 13 kinases of the AMPK subfamily, including MARK/PAR-1. EMBO J. 23, 833–843 (2004).
Corradetti, M.N., Inoki, K., Bardeesy, N., DePinho, R.A. & Guan, K.-L. Regulation of the TSC pathway by LKB1: evidence of a molecular link between tuberous sclerosis complex and Peutz-Jeghers syndrome. Genes Dev. 18, 1533–1538 (2004).
Resta, N. et al. Two novel mutations and a new STK11/LKB1 gene isoform in Peutz-Jeghers patients. Hum. Mutat. 20, 78–79 (2002).
Hastings, M.L. & Krainer, A.R. Splicing in the new millennium. Curr. Opin. Cell Biol. 13, 302–309 (2001).
Patel, A.A. & Steitz, J.A. Splicing double: insights from the second spliceosome. Nat. Rev. Mol. Cell Biol. 4, 960–970 (2003).
Levine, A. & Durbin, R. A computational scan for U12-dependent introns in the human genome sequence. Nucleic Acids Res. 29, 4006–4013 (2001).
Burset, M., Seledtsov, I.A. & Solovyev, V.V. Analysis of canonical and non-canonical splice sites in mammalian genomes. Nucleic Acids Res. 28, 4364–4375 (2000).
Maquat, L.E. Nonsense-mediated mRNA decay: splicing, translation and mRNP dynamics. Nat. Rev. Mol. Cell Biol. 5, 89–99 (2004).
Nakai, K. & Sakamoto H. Construction of a novel database containing aberrant splicing mutations of mammalian genes. Gene 141, 171–177 (1994).
Roca, X., Sachidanandam, R. & Krainer, A.R. Intrinsic differences between authentic and cryptic 5′ splice sites. Nucleic Acids Res. 31, 6321–6333 (2003).
Hastings, M.L. & Krainer, A.R. Functions of SR proteins in the U12-dependent AT-AC pre-mRNA splicing pathway. RNA 7, 471–482 (2001).
Senapathy, P., Shapiro, M.B. & Harris, N.L. Splice junctions, branch point sites, and exons: sequence statistics, identification, and applications to genome project. Methods Enzymol. 183, 252–278 (1990).
Krainer, A.R., Maniatis, T., Ruskin, B. & Green, M.R. Normal and mutant human β-globin pre-mRNAs are faithfully and efficiently spliced in vitro. Cell 36, 993–1005 (1984).
Frilander, M.J. & Steitz, J.A. Initial recognition of U12-dependent introns requires both U11/5′ splice site and U12/branchpoint interactions. Genes Dev. 13, 851–863 (1999).
Will, C.L. et al. The human 18S U11/U12 snRNP contains a set of novel proteins not found in the U2-dependent spliceosome. RNA 10, 929–941 (2004).
McConnell, T.S., Cho, S.-J., Frilander, M.J. & Steitz, J.A. Branchpoint selection in the splicing of U12-dependent introns in vitro. RNA 8, 579–586 (2002).
Dietrich, R.C., Incorvaia, R. & Padgett, R.A. Terminal intron dinucleotide sequences do not distinguish between U2- and U12- dependent introns. Mol. Cell 1, 151–160 (1997).
Dietrich, R.C., Peris, M.J., Seyboldt, A.S. & Padgett, R.A. Role of the 3′ splice site in U12-dependent intron splicing. Mol. Cell. Biol. 21, 1942–1952 (2001).
Collins, C.A. & Guthrie, C. Allele-specific genetic interactions between Prp8 and RNA active site residues suggest a function for Prp8 at the catalytic core of the spliceosome. Genes Dev. 13, 1970–1982 (1999).
Siatecka, M., Reyes, J.L., & Konarska, M.M. Functional interactions of Prp8 with both splice sites at the spliceosomal catalytic center. Genes Dev. 13, 1983–1993 (1999).
Luo, H.R., Moreau, G.A., Levin, N. & Moore, M.J. The human Prp8 protein is a component of both U2- and U12-dependent spliceosomes. RNA 5, 8893–8908 (1999).
Wu, Q. & Krainer, A.R. AT-AC pre-mRNA splicing mechanisms and conservation of minor introns in voltage-gated ion channel genes. Mol. Cell. Biol. 19, 3225–3236 (1999).
Burge, C.B., Padgett, R.A. & Sharp P.A. Evolutionary fates and origins of U12-type introns. Mol. Cell 2, 773–785 (1998).
Clark, F. & Thanaraj, T.A. Categorization and characterization of transcript-confirmed constitutively and alternatively spliced introns and exons from human. Hum. Mol. Gen. 11, 451–464 (2002).
Otake, L.R., Scamborova, P., Hashimoto, C. & Steitz, J.A. The divergent U12-type spliceosome is required for pre-mRNA splicing and is essential for development in Drosophila. Mol. Cell 9, 439–446 (2002).
Kohrman, D.C., Harris, J.B. & Meisler, M.H. Mutation detection in the med and medJ alleles of the sodium channel Scn8a. J. Biol. Chem. 271, 17576–17581 (1996).
Shaw, M.A. et al. Identification of three novel SEDL mutations, including mutation in the rare, non-canonical splice site of exon 4. Clin. Genet. 64, 235–242 (2003).
Acknowledgements
This work was supported by US National Institutes of Health grant GM42699 (A.R.K.) and MIUR-FIRB RBAU01SZHB-001 (G.G.). M.L.H. was supported by an American Cancer Society postdoctoral fellowship. We thank L. Manche and A. Quagliarella for technical assistance.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Hastings, M., Resta, N., Traum, D. et al. An LKB1 AT-AC intron mutation causes Peutz-Jeghers syndrome via splicing at noncanonical cryptic splice sites. Nat Struct Mol Biol 12, 54–59 (2005). https://doi.org/10.1038/nsmb873
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb873
This article is cited by
-
Defective minor spliceosomes induce SMA-associated phenotypes through sensitive intron-containing neural genes in Drosophila
Nature Communications (2020)
-
Characterization of the STK11 splicing variant as a normal splicing isomer in a patient with Peutz–Jeghers syndrome harboring genomic deletion of the STK11 gene
Human Genome Variation (2016)
-
A Candidate Gene Association Study for Growth Performance in an Improved Giant Freshwater Prawn (Macrobrachium rosenbergii) Culture Line
Marine Biotechnology (2014)
-
DAAM2 polymorphism is closely related to the clinical outcomes of allogeneic hematopoietic stem cell transplantation
Annals of Hematology (2012)
-
A comparative analysis of exome capture
Genome Biology (2011)