Abstract
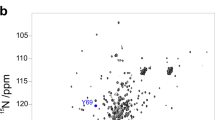
The modern view of protein thermodynamics predicts that proteins undergo cold-induced unfolding. Unfortunately, the properties of proteins and water conspire to prevent the detailed observation of this fundamental process. Here we use protein encapsulation to allow cold denaturation of the protein ubiquitin to be monitored by high-resolution NMR at temperatures approaching −35 °C. The cold-induced unfolding of ubiquitin is found to be highly noncooperative, in distinct contrast to the thermal melting of this and other proteins. These results demonstrate the potential of cold denaturation as a means to dissect the cooperative substructures of proteins and to provide a rigorous framework for testing statistical thermodynamic treatments of protein stability, dynamics and function.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$189.00 per year
only $15.75 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Accession codes
Change history
07 March 2004
Added note and updated figure online, updated PDF
Notes
*Note: In the version of this article initially published online, the temperature for the far right-hand panels of Figure 3a,b was incorrectly labeled during the production process. The correct temperature should be −30 °C. We apologize for any inconvenience this may have caused. This mistake has been corrected for the HTML and print versions of the article.
References
Privalov, P.L. Cold denaturation of proteins. Crit. Rev. Biochem. Mol. Biol. 25, 281–305 (1990).
Nash, D.P. & Jonas, J. Structure of the pressure-assisted cold denatured state of ubiquitin. Biochem. Biophys. Res. Comm. 238, 289–291 (1997).
Bai, Y., Sosnick, T.R., Mayne, L. & Englander, S.W. Protein folding intermediates: native-state hydrogen exchange. Science 269, 192–197 (1995).
Fuentes, E.J. & Wand, A.J. Local stability and dynamics of apocytochrome b562 examined by the dependence of hydrogen exchange on hydrostatic pressure. Biochemistry 37, 9877–9883 (1998).
Hatley, R.H.M. & Franks, F. The cold-induced denaturation of lactate dehydrogenase at sub-zero temperatures in the absence of perturbants. FEBS Lett. 257, 171–173 (1989).
Wand, A.J., Ehrhardt, M.R. & Flynn, P.F. High-resolution NMR of encapsulated proteins dissolved in low-viscosity fluids. Proc. Natl. Acad. Sci. USA 95, 15299–15302 (1998).
Babu, C.R., Flynn, P.F. & Wand, A.J. Validation of protein structure from preparations of encapsulated proteins dissolved in low viscosity fluids. J. Am. Chem. Soc. 123, 2691–2692 (2001).
Wintrode, P.L., Makhatadze, G.I. & Privalov, P.L. Thermodynamics of ubiquitin unfolding. Proteins 18, 246–253 (1994).
Ibarra-Molero, B., Makhatadze, G.I. & Sanchez-Ruiz, J.M. Cold denaturation of ubi-quitin. Biochim. Biophys. Acta 1429, 384–390 (1999).
Skalicky, J.J., Sukumaran, D.K., Mills, J.L. & Szyperski, T. Toward structural biology in supercooled water. J. Am. Chem. Soc. 122, 3230–3231 (2000).
Zhou, H.X. & Dill, K.A. Stabilization of proteins in confined spaces. Biochemistry 40, 11289–11293 (2001).
Shastry, M.C.R. & Eftink, M.R. Reversible thermal unfolding of ribonuclease T1 in reverse micelles. Biochemistry 35, 4094–4101 (1996).
Battistel, E., Luisi, P.L. & Rialdi, G. Thermodynamic study of globular protein stability in microemulsions. J. Phys. Chem. 92, 6680–6685 (1988).
Dill, K.A., Fiebig, K.M. & Chan, H.S. Cooperativity in protein-folding kinetics. Proc. Natl. Acad. Sci. USA 90, 1942–1946 (1993).
Sivaraman, T., Arrington, C.B. & Robertson, A.D. Kinetics of unfolding and folding from amide hydrogen exchange in native ubiquitin. Nat. Struct. Biol. 8, 331–331 (2001).
Briggs, M.S. & Roder, H. Proc. Early hydrogen-bonding events in the folding reaction of ubiquitin. Proc. Natl. Acad. Sci. USA 89, 2017–2021 (1992).
Shortle, D. & Ackerman, M.S. Persistence of native-like topology in a denatured protein in 8 M urea. Science 293, 487–489 (2001)
Hilser, V.J. & Freire, E. Structure-based calculation of the equilibrium folding pathway of proteins. Correlation with hydrogen exchange protection factors. J. Mol. Biol. 262, 756–772 (1996).
Hilser, V.J., Dowdy, D., Oas, T. & Freire, E. The structural distribution of cooperative interactions in proteins: analysis of the native state ensemble. Proc. Natl. Acad. Sci. USA 95, 9903–9908 (1998).
Ooi, T. & Oobatake, M. Prediction of the thermodynamics of protein unfolding: the helix-coil transition of poly(L-alanine). Proc. Natl. Acad. Sci. USA 88, 2859–2863 (1991).
Wand, A.J., Urbauer, J.L., McEvoy, R.P. & Bieber, R.J. Internal dynamics of human ubiquitin revealed by 13C-relaxation of randomly, fractionally enriched protein. Biochemistry 35, 6116–6125 (1996).
Acknowledgements
This work was supported by grants from the US National Institutes of Health. We are grateful to K.A. Sharp and S.W. Englander for helpful discussion and to S. Whitten and P.F. Flynn for technical assistance.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Babu, C., Hilser, V. & Wand, A. Direct access to the cooperative substructure of proteins and the protein ensemble via cold denaturation. Nat Struct Mol Biol 11, 352–357 (2004). https://doi.org/10.1038/nsmb739
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsmb739
This article is cited by
-
A review on protein oligomerization process
International Journal of Precision Engineering and Manufacturing (2015)
-
Cold denaturation of a protein dimer monitored at atomic resolution
Nature Chemical Biology (2013)
-
Key stabilizing elements of protein structure identified through pressure and temperature perturbation of its hydrogen bond network
Nature Chemistry (2012)
-
Quantitative Assessment of Protein Structural Models by Comparison of H/D Exchange MS Data with Exchange Behavior Accurately Predicted by DXCOREX
Journal of the American Society for Mass Spectrometry (2012)
-
Site-resolved measurement of water-protein interactions by solution NMR
Nature Structural & Molecular Biology (2011)