Abstract
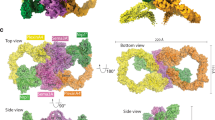
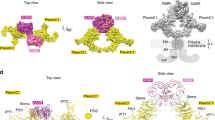
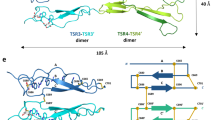
Semaphorins, proteins characterized by an extracellular sema domain, regulate axon guidance, immune function and angiogenesis. The crystal structure of SEMA4D (residues 1–657) shows the sema topology to be a seven-bladed β-propeller, revealing an unexpected homology with integrins. The sema β-propeller contains a distinctive 77-residue insertion between β-strands C and D of blade 5. Blade 7 is followed by a domain common to plexins, semaphorins and integrins (PSI domain), which forms a compact cysteine knot abutting the side of the propeller, and an Ig-like domain. The top face of the β-propeller presents prominent loops characteristic of semaphorins. In addition to limited contact between the Ig-like domains, the homodimer is stabilized through extensive interactions between the top faces in a sector of the β-propeller used for heterodimerization in integrins. This face of the propeller also mediates ligand binding in integrins, and functional data for semaphorin-receptor interactions map to the equivalent surface.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Accession codes
References
Kolodkin, A.L., Matthes, D.J. & Goodman, C.S. The semaphorin genes encode a family of transmembrane and secreted growth cone guidance molecules. Cell 75, 1389–1399 (1993).
Semaphorin Nomenclature Committee (S.N.). Unified nomenclature for the semaphorins/collapsins. Cell 97, 551–552 (1999).
Luo, Y., Raible, D. & Raper, J.A. Collapsin: a protein in brain that induces the collapse and paralysis of neuronal growth cones. Cell 75, 217–227 (1993).
Raper, J.A. Semaphorins and their receptors in vertebrates and invertebrates. Curr. Opin. Neurobiol. 10, 88–94 (2000).
Takahashi, T. et al. Plexin–neuropilin-1 complexes form functional semaphorin-3A receptors. Cell 99, 59–69 (1999).
Tamagnone, L. et al. Plexins are a large family of receptors for transmembrane, secreted, and GPI-anchored semaphorins in vertebrates. Cell 99, 71–80 (1999).
He, Z. & Tessier-Lavigne, M. Neuropilin is a receptor for the axonal chemorepellent Semaphorin III. Cell 90, 739–751 (1997).
Kolodkin, A.L. et al. Neuropilin is a semaphorin III receptor. Cell 90, 753–762 (1997).
Winberg, M.L. et al. Plexin A is a neuronal semaphorin receptor that controls axon guidance. Cell 95, 903–916 (1998).
Bork, P., Doerks, T., Springer, T.A. & Snel, B. Domains in plexins: links to integrins and transcription factors. Trends Biochem. Sci. 24, 261–263 (1999).
Xiong, J.P. et al. Crystal structure of the extracellular segment of integrin αVβ3. Science 294, 339–345 (2001).
Hall, K.T. et al. Human CD100, a novel leukocyte semaphorin that promotes B-cell aggregation and differentiation. Proc. Natl. Acad. Sci. USA 93, 11780–11785 (1996).
Shi, W. et al. The class IV semaphorin CD100 plays nonredundant roles in the immune system: defective B and T cell activation in CD100-deficient mice. Immunity 13, 633–642 (2000).
Kumanogoh, A. et al. Identification of CD72 as a lymphocyte receptor for the class IV semaphorin CD100: a novel mechanism for regulating B cell signaling. Immunity 13, 621–631 (2000).
Giordano, S. et al. The semaphorin 4D receptor controls invasive growth by coupling with Met. Nat. Cell. Biol. 4, 720–724 (2002).
Sprague, E.R., Redd, M.J., Johnson, A.D. & Wolberger, C. Structure of the C-terminal domain of Tup1, a corepressor of transcription in yeast. EMBO J. 19, 3016–3027 (2000).
Wall, M.A. et al. The structure of the G protein heterotrimer Gi α1 β1 γ2. Cell 83, 1047–1058 (1995).
Sondek, J., Bohm, A., Lambright, D.G., Hamm, H.E. & Sigler, P.B. Crystal structure of a G-protein βγ dimer at 2.1 Å resolution. Nature 379, 369–374 (1996).
Stuart, D.I., Levine, M., Muirhead, H. & Stammers, D.K. Crystal structure of cat muscle pyruvate kinase at a resolution of 2.6 Å. J. Mol. Biol. 134, 109–142 (1979).
Calvete, J.J., Henschen, A. & Gonzalez-Rodriguez, J. Assignment of disulphide bonds in human platelet GPIIIa. A disulphide pattern for the β-subunits of the integrin family. Biochem. J. 274, 63–71 (1991).
Behar, O., Mizuno, K., Badminton, M. & Woolf, C.J. Semaphorin 3A growth cone collapse requires a sequence homologous to tarantula hanatoxin. Proc. Natl. Acad. Sci. USA 96, 13501–13505 (1999).
Klostermann, A., Lohrum, M., Adams, R.H. & Puschel, A.W. The chemorepulsive activity of the axonal guidance signal semaphorin D requires dimerization. J. Biol. Chem. 273, 7326–7331 (1998).
Koppel, A.M. & Raper, J.A. Collapsin-1 covalently dimerizes, and dimerization is necessary for collapsing activity. J. Biol. Chem. 273, 15708–15713 (1998).
Koppel, A.M., Feiner, L., Kobayashi, H. & Raper, J.A. A 70 amino acid region within the semaphorin domain activates specific cellular response of semaphorin family members. Neuron 19, 531–537 (1997).
Russell, R.B., Sasieni, P.D. & Sternberg, M.J. Supersites within superfolds. Binding site similarity in the absence of homology. J. Mol. Biol. 282, 903–918 (1998).
Pasterkamp, R.J. & Kolodkin, A.L. Semaphorin junction: making tracks toward neural connectivity. Curr. Opin. Neurobiol. 13, 79–89 (2003).
Kamata, T. & Takada, Y. Platelet integrin αIIbβ3–ligand interactions: what can we learn from the structure? Int. J. Hematol. 74, 382–389 (2001).
Xiong, J.P. et al. Crystal structure of the extracellular segment of integrin α Vβ3 in complex with an Arg-Gly-Asp ligand. Science 296, 151–155 (2002).
Davis, S.J. et al. High level expression in Chinese hamster ovary cells of soluble forms of CD4 T lymphocyte glycoprotein including glycosylation variants. J. Biol. Chem. 265, 10410–10418 (1990).
Stanley, P. Selection of specific wheat germ agglutinin-resistant (WgaR) phenotypes from Chinese hamster ovary cell populations containing numerous lecR genotypes. Mol. Cell Biol. 1, 687–696 (1981).
May, A.P. et al. Expression, crystallization, and preliminary X-ray analysis of a sialic acid-binding fragment of sialoadhesin in the presence and absence of ligand. Protein Sci. 6, 717–721 (1997).
Davis, S.J. et al. Crystallization and functional analysis of a soluble deglycosylated form of the human costimulatory molecule B7-1. Acta Crystallogr. D 57, 605–608 (2001).
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997).
Weeks, C.M. & Miller, R. The design and implementation of SnB version 2.0. J. Appl. Crystallogr. 32, 120–124 (1999).
Brunger, A.T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998).
Schneider, T.R. & Sheldrick, G.M. Substructure solution with SHELXD. Acta Crystallogr. D 58, 1772–1779 (2002).
Terwilliger, T.C. Automated main-chain model building by template matching and iterative fragment extension. Acta Crystallogr. D 59, 38–44 (2003).
Terwilliger, T.C. Automated side-chain model building and sequence assignment by template matching. Acta Crystallogr. D 59, 45–49 (2003).
Jones, T.A., Zou, J.Y., Cowan, S.W. & Kjeldgaard, M. Improved methods of the building of proteins models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991).
Brunger, A.T. X-PLOR. Version 3.1. A System for X-ray Crystallography and NMR (Yale University Press, New Haven, Connecticut, USA, 1992).
Cowtan, K.D. & Zhang, K.Y. Density modification for macromolecular phase improvement. Prog. Biophys. Mol. Biol. 72, 245–270 (1999).
Morris, R.J., Perrakis, A. & Lamzin, V.S. ARP/wARP's model-building algorithms. I. The main chain. Acta Crystallogr. D 58, 968–975 (2002).
Navaza, J. Implementation of molecular replacement in AMoRe. Acta Crystallogr. D 57, 1367–1372 (2001).
Esnouf, R.M. Further additions to MolScript version 1.4, including reading and contouring of electron-density maps. Acta Crystallogr. D 55, 938–940 (1999).
Tomizawa, Y. et al. Inhibition of lung cancer cell growth and induction of apoptosis after reexpression of 3p21.3 candidate tumor suppressor gene SEMA3B. Proc. Natl. Acad. Sci. USA 98, 13954–13959 (2001).
Xiang, R.H. et al. Isolation of the human semaphorin III/F gene (SEMA3F) at chromosome 3p21, a region deleted in lung cancer. Genomics 32, 39–48 (1996).
Acknowledgements
We are grateful to R. Manso-Sancho for advice and assistance in protein expression, L. Lyne and W. Lu for cell culture, A. Chedatol and L. Tamagnone for biological assays of sSEMA4D, T. Batuwangala for additional work on semaphorins, N. Zaccai for data collection, J. Brown, J. Grimes and P. Roche for help with computation and S. Naylor for discussion. We thank M. Walsh and colleagues at BM14 as well as the staff of the ESRF and EMBL outstation in Grenoble. This work was funded by Cancer Research UK with additional support from the European Commission Integrated Program SPINE. D.I.S. and E.Y.J. are supported by the UK Medical Research Council and Cancer Research UK, respectively.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Love, C., Harlos, K., Mavaddat, N. et al. The ligand-binding face of the semaphorins revealed by the high-resolution crystal structure of SEMA4D. Nat Struct Mol Biol 10, 843–848 (2003). https://doi.org/10.1038/nsb977
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nsb977
This article is cited by
-
Diversity of oligomerization in Drosophila semaphorins suggests a mechanism of functional fine-tuning
Nature Communications (2019)
-
Neuroimmune semaphorins as costimulatory molecules and beyond
Molecular Medicine (2018)
-
Variable expressivity and genetic heterogeneity involving DPT and SEMA3D genes in autosomal dominant familial Meniere’s disease
European Journal of Human Genetics (2017)
-
Monitoring storage induced changes in the platelet proteome employing label free quantitative mass spectrometry
Scientific Reports (2017)
-
Viral glycoproteins: biological role and application in diagnosis
VirusDisease (2016)