Key Points
-
Chaperone–usher (CU) pili are assembled at the outer membrane by two proteins, a periplasmic chaperone, which provides the scaffold for the correct folding of pilins, and an outer-membrane protein called the usher, which forms a dimeric complex at the outer membrane to recruit and polymerize chaperone–subunit complexes and translocate the growing pilus to the outer surface. CU pili comprise linear, unbranched polymers of several hundreds to thousands of 12–20 kDa pilin subunits. This Review focuses on the rod-like fimbrial organelles, particularly uropathogenic Escherichia coli P and type 1 pili (the Pap and Fim systems, respectively).
-
Over the past decade, a plethora of structural information has been gathered about the various players that participate in P and type 1 pilus biogenesis, and these structural data, including the newly determined usher structures, are reviewed in detail.
-
Pilin subunits contain an incomplete, immunoglobulin-like fold that lacks the carboxy-terminal β-strand, which results in the presence of a large hydrophobic groove. When the chaperone–pilin complex is formed, a structural motif on the G1 strand of the chaperone is inserted into this groove. This donor strand complementation reaction is discussed in light of the most recent structural and biochemical data.
-
Pilin subunits also contain an amino-terminal extension (Nte) peptide that is disordered in the chaperone–subunit complex. During polymerization, the Nte on the incoming pilin subunit replaces the chaperone G1 strand, and this donor strand exchange reaction, including the 'zip-in, zip-out' mechanism, is also reviewed.
-
Recent structural data have indicated that only a single usher pore in the dimeric usher complex is used for pilus secretion. This model is discussed in detail.
-
Bacterial attachment to mucosal surfaces can be the result of the specific recognition of a pilus-associated adhesin by a host cell receptor. The molecular basis of receptor recognition of the FimH and PapG adhesins is described.
-
Finally, the complex role of CU pili in the pathogenesis of urinary tract infections is summarized, along with the prospects for using CU pili as a target for novel antibacterial therapies.
Abstract
The chaperone–usher (CU) pathway of pilus biogenesis is the most widespread of the five pathways that assemble adhesive pili at the surface of Gram-negative bacteria. Recent progress in the study of the structural biology of the CU pathway has unravelled the molecular basis of chaperone function and elucidated the mechanisms of fibre assembly at the outer membrane, leading to a comprehensive description of each step in the biogenesis pathway. Other studies have provided the molecular basis of host recognition by CU pili. The knowledge that has been gathered about both the assembly of and host recognition by CU pili has been harnessed to design promising antibiotic compounds.
Similar content being viewed by others
Main
Bacteria commonly express proteinaceous appendages on their outer surfaces. One class of extracellular polymers, known as pili or fimbriae, is used in attachment and invasion, biofilm formation, cell motility and transport of proteins and DNA across membranes. These non-flagellar appendages of Gram-negative bacteria can be categorized into five major classes on the basis of the biosynthetic pathway involved: chaperone–usher pili (CU pili), curli, type IV pili, the type III secretion needle and type IV secretion pili (reviewed in Ref. 1). Of these five classes, the CU pili are the most extensively studied. CU pili are assembled at the outer membrane by two proteins, a periplasmic chaperone and an outer-membrane, pore-forming protein called the usher2. The chaperone facilitates folding of pilus subunits3, prevents them from polymerizing in the periplasm and targets them to the usher4,5. The usher acts as an assembly platform, recruiting chaperone–subunit complexes from the periplasm, coordinating their assembly into a pilus and secreting that pilus through the usher pore. Investigations of the structure of CU pili have elucidated not only the mechanism of pilus assembly but also the functions of these pili in host–pathogen interactions. Here, we review the spectacular progress that has taken place over the past 10 years and has led to an overall understanding of the biogenesis and function of CU pili.
CU pilus morphology and structure
CU pili are assembled into linear, unbranched polymers consisting of several hundreds to thousands of pilus subunits (also known as pilins) that range in size from ∼12 kDa to ∼20 kDa. CU organelles differ widely in complexity and morphology, ranging from thin, fibrillar fibres to thick rods composed of a helically wound cylinder and a distinct fibrillar tip. Phylogenetic analysis of usher sequences in all 189 known CU systems revealed 6 main clades: α-, β-, γ- (which is subdivided into γ1, γ2, γ3 and γ4), κ-, π- and σ-fimbriae6. This Review focuses on the rod-like or 'typical' fimbrial organelles, which are found in the α-, γ- and π-fimbrial clades. Among these, the P and type 1 pili (belonging to the π- and γ1 fimbrial clades, respectively) of uropathogenic Escherichia coli (UPEC) (Box 1) have been the focus of extensive studies.
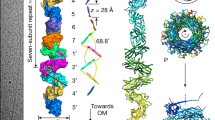
P pili consist of six different subunit types arranged into two distinct subassemblies, the tip fibrillum and the pilus rod. The flexible, distal tip fibrillum is ∼2 nm in diameter and is composed of one copy of the PapG adhesin at the distal end, followed by one copy of the adaptor subunit PapF and 5–10 copies of the PapE subunit (Fig. 1). The long, rigid, 6.8 nm-wide pilus rod is composed of more than 1,000 copies of the PapA subunit7 (Fig. 1), which are connected to the tip fibrillum by one copy of the adaptor subunit PapK and terminated at the proximal end (in the cell wall) by one copy of the termination subunit PapH. The PapA rod adopts a right-handed, one-start superhelical structure with a 2.5 nm pitch and 3.3 subunits per turn8,9. Type 1 pili display a similar but simplified architecture, with shorter tip fibrillae10 consisting of one copy each of FimH (the adhesin), FimG and FimF (Fig. 1). The major rod subunit is FimA. No termination subunit has been identified in the type 1 pilus to date.
A schematic of P (part a) and type 1 (part b) pili, represented by the Pap and Fim systems, respectively. Numbers indicate the number of copies of each subunit in the pilus. The chaperones attached to the last subunit to be incorporated into each pilus are shown in yellow. P pili are terminated at the outer membrane (OM) by the termination subunit, PapH. No such subunit is known in the Fim system. The usher dimers are indicated in purple and blue. E, extracellular space; P, periplasm. Figure is modified, with permission, from Ref. 35 © (2008) Elsevier.
Pilus assembly
Chaperone–subunit interactions. Pilus subunits are taken up by their cognate periplasmic chaperones as soon as they exit the Sec machinery (which mediates general secretion)11. In the absence of the chaperone, the subunits cannot fold properly and form aggregates that are targeted to the DegP protease for degradation3,12,13. Thus, chaperones provide the scaffold onto and around which pilus subunits fold. The periplasmic chaperones are 'boomerang'-shaped, ∼25 kDa proteins consisting of two immunoglobulin-like (Ig-like) domains14,15. The interactive surfaces that contact the pilus subunit are formed by two conserved basic residues located between the two Ig-like domains (specifically, Arg8 and Lys112, following Pap numbering)16 and by the amino-terminal and seventh β-strands (strands A1 and G1, respectively) of the N-terminal domain of the chaperone (Fig. 2a,b). Mutations of Arg8 or Lys112 in the Pap system completely abrogate pilus biogenesis15. Pilus subunits are characterized by an incomplete Ig-like fold that lacks the C-terminal β-strand17,18 (Fig. 2b). As a result, all subunits possess a large groove where the missing strand would normally be in a complete Ig fold. In chaperone–subunit complexes, a motif of four alternating hydrophobic residues on the chaperone G1 strand are inserted into this hydrophobic groove. These four residues are designated P1 to P4, and their corresponding acceptor sites in the subunit groove are known as the P1 to P4 pockets or sites (Fig. 2a). The interaction between the subunit groove and the chaperone G1 strand also includes the formation of hydrogen bonds between the main-chain atoms of the groove-flanking β-strands of the subunit and those of the chaperone G1 strand. Thus, the chaperone provides in trans (or 'donates') the secondary structural element that is missing from the pilus subunit, a process termed donor strand complementation (DSC)17,18 (Fig. 2). By providing the structural information that is missing from the subunit Ig-like fold, the chaperone functions as a structural platform that catalyses the correct folding of the pilus subunits3,13,19.
a | A ribbon diagram of the PapD–PapK complex structure. PapD and PapK are in green and cyan, respectively. The carboxyl terminus of PapK is shown as a ball-and-stick representation. The conserved chaperone residues that contactthe subunit C terminus (Arg8 and Lys112) and the P2, P3 and P4 residues in the chaperone G1 β-strand are shown in the same representation. b | A topology diagram of PapK. Arrows and cylinders represent β-strands and α-helices, respectively. The C and amino termini are indicated. Note that in the chaperone–subunit complex structure, the N-terminal extension (Nte) is disordered and thus not represented in the topology diagram of the subunit prior to donor strand exchange (DSE). The red arrow represents the G1 β-strand donated by the chaperone. c | The structure of the pilus subunit after DSE. The subunit is shown in a ribbon diagram (cyan). The donor strand (or Nte) from the incoming subunit is shown in red. The P2, P3, P4 and P5 residues in the donor strand are shown in a ball-and-stick representation (red). d | A topological diagram of the subunits in the pilus. The representation is as in part b. The Nte (red) of the following subunit complements in trans the fold of the previously assembled subunit. e | A close-up of the P5 pocket (circled in green) and P4 pocket of PapA. f | A sequence alignment of the Ntes of all Pap subunits (except for PapG, which does not have an Nte). The alternating hydrophobic residue motif (DSE region) is highlighted in yellow, with the conserved Gly in magenta. Parts b and d are modified, with permission, from Ref. 35 © (2008) Elsevier. Part e is reproduced, with permission, from Ref. 31 © (2008) Elsevier.
Depending on the length of the loop between the F1 and G1 strands (the F1G1 loop) of the chaperone, two structural scaffolds are observed20. In chaperone–subunitinteractions that involve chaperones with short F1G1 loops, termed FGS chaperones, the G1 donor strand from the chaperone inserts between the A and F strands of the pilus subunit to form a continuous β-sheet17,18 (Fig. 2a). In interactions that involve chaperones with long F1G1 loops, termed FGL chaperones, the A and F strands of the pilus subunit form the edge strands of the two opposing sheets in the Ig-like β-sandwich21,22,23. FGL chaperones contain an additional alternating hydrophobic residue on the G1 strand (known as P5, as it follows P4) that inserts intermittently into a corresponding P5 pocket in the subunit groove. In subunits that interact with FGS chaperones, this P5 pocket is never occupied by a chaperone residue, as the G1 strand is too short.
In all chaperone–subunit interactions, the donated chaperone G1 strand runs parallel to strand F in the subunit, resulting in an atypical Ig fold: in a typical Ig fold, the seventh strand (strand G) runs antiparallel to strand F. The consequence of this atypical fold complementation event is that the subunit is maintained in a polymerization-competent state, which is characterized by the accessibility of the P5 pocket to incoming subunits during pilus assembly and a partially flexible conformation in which a number of loops and secondary structural elements in the chaperone-bound subunit are disordered.
Subunit–subunit interactions. All CU pilus subunits contain a 10–20 residue-long N-terminal extension (Nte) peptide that is disordered in the chaperone–subunit complex and is not part of the subunit fold. During subunit polymerization, the complementing G1 β-strand donated by the chaperone is replaced by the Nte on the subunit of the incoming chaperone–subunit complex. This assembly reaction is termed donor strand exchange (DSE)21,22,24 (Fig. 2c,d). Nte sequences are relatively conserved, as they contain a set of alternating hydrophobic residues that form an essential part of the subunit–subunit interaction. The P1 to P4 pockets in the subunit groove are occupied by the P1 to P4 residues of the chaperone G1 strand (discussed above) in the chaperone–subunit complex, but after DSE it is the P2 to P5 pockets of the subunit groove that are occupied by the hydrophobic residues (termed P2–P5 residues) of the incoming subunit Nte (Fig. 2a,c,f). The P4 Gly residue in Nte sequences is strictly conserved (Fig. 2f). This residue lies on top of the P4 pocket in the subunit groove, which contains a bulk that is usually formed by an aromatic residue (Fig. 2e). Gly is the only residue that can accommodate this bulk, as any other amino acid would give rise to steric constraints that would destabilize the interaction between the groove and the Nte. Thus, the interaction of P4 Gly with the bulk of the P4 pocket ensures that the incoming Nte registers correctly in the accepting groove.
The DSE 'zip-in, zip-out' mechanism. The exchange reaction between the chaperone G1 strand and the Nte in the incoming subunit occurs through a concerted 'zip-in, zip-out' mechanism that is initiated by the insertion of the Nte P5 residue into the P5 pocket of the groove in the previously assembled subunit22,25 (Fig. 3a). The identification of a ternary chaperone–subunit Nte complex during in vitro DSE reactions in Salmonella SafA subunits, using non-denaturing mass spectrometry, was crucial to the discovery of this mechanism22. The formation of the ternary complex was shown to depend on the accessibility of the P5 pocket in the accepting pilus subunit (Fig. 3b). As mentioned above, for pilus subunits that are assembled by FGS chaperones, the P5 pocket is not occupied by the chaperone G1 strand and is therefore freely accessible to the P5 residue of the Nte of an incoming pilus subunit. In systems such as the Salmonella Saf pili, which are assembled by FGL chaperones, the chaperone occupies the P5 site but does so in an equilibrium with a locally unbound, accessible state22. In the Saf system, mutating the chaperone G1 residue that occupies the P5 site to a residue with an improved steric fit to the pocket (an Ala to Phe substitution, in the case of theSaf system) shifts the equilibrium towards the closed state, thus suppressing formation of the ternary complex and drastically reducing the DSE rate. Similarly, decreasing the hydrophobic bulk of the P5 residue on the Nte of the incoming subunit negatively affects ternary complex formation and the DSE rate. Thus, the P5 pocket in a chaperone-bound subunit forms the primary acceptor site for the Nte of an incoming subunit. The interaction between the Nte (through its P5 residue) and the P5 pocket initiates DSE. Single-site mutagenesis of the P5, P4 and P3 residues in the Nte to Ala revealed a gradient of decreasing DSE efficiency moving away from the P5 initiation site, suggesting a 'zip-in, zip-out' mechanism, with the new Nte gradually displacing the chaperone G1 donor strand in a step-wise process from P5 to P1 (Ref. 22) (Fig. 3a).
a | A schematic diagram for donor strand exchange (DSE). Progressive insertion of the P5, P4, P3 and P2 residues of the attacking amino-terminal extension (Nte) is shown in pink. b | The presence of a P5 pocket in PapK (left panel) and the absence of a corresponding pocket in PapH (right panel). All Pap subunits except PapH have a P5 pocket, which serves as an initiation site for the progressive zipping-in of the Nte of the subunit that comes next in assembly. As PapH does not have a P5 pocket, an Nte cannot challenge the PapD–PapH complex. Part b is reproduced, with permission, from EMBO Reports Ref. 9 © (2006) Macmillan Publishers Ltd. All rights reserved.
Termination of pilus biogenesis. Understanding the crucial role of the P5 pocket in DSE also helps us to understand the molecular basis of the termination of pilus biogenesis. In the P pilus system, termination occurs when the PapD–PapH chaperone–subunit complex is presented for assembly at the usher8. PapH undergoes DSE with the last PapA subunit of the pilus rod. However, PapH is unable to undergo DSE with any other subunits, as confirmed by in vitro experiments in which the PapD–PapH complex was refractory to challenge by Nte peptides derived from any of the Pap subunits9. This restriction is not due to a stronger chaperone–subunit interaction but has been ascribed instead to a single distinguishing structural feature: the absence of a P5 pocket in PapH9 (Fig. 3b). The mechanism by which type 1 pilus biogenesis terminates is not known, as no homologue of PapH has been found in that system to date.
Subunit ordering. The location of PapG at the tip of the P pilus was first discovered by Lindberg et al.26. Later, high-resolution electron microscopy (EM) studies on various P pilus mutants elucidated the composite nature of the pilus and the fact that PapE constitutes the bulk of the tip fibrillum7. These studies were subsequently extended to deduce the order of the subunits in the pilus: PapF links the adhesin, PapG, to the tip fibrillum polymer, PapE, and PapK links the tip fibrillum to the pilus rod27. It was then shown that PapA and PapE are the only subunits in the pilus that have self-associating properties28, consistent with the results from genetic and EM studies. The role of Ntes in specifying subunit–subunit interactions was first demonstrated by genetic and biochemical experiments29 that used a series of N-terminally swapped subunits. PapF, which is required to link PapG to PapE27, was proposed to have an Nte that fits into the PapG pilin groove. The Nte of PapF was swapped onto the pilin body of PapE to produce a fusion protein that could complement ΔpapF mutants and link PapG to the PapE tip fibrillum29. Cells encoding a PapF that was missing the Nte behaved in a similar manner to ΔpapF mutants and were unable to incorporate PapG into the pilus. The results that were obtained using various combinations of N-terminally swapped and deleted subunits support the proposal that there is a role for Ntes in specifying subunit order in the pilus.
The role of the Nte–groove interaction in determining subunit ordering was confirmed by a study that examined the specificity of DSE in Pap subunit assembly30. An in vitro DSE assay was set up, in which all six chaperone–subunit complexes of the Pap system were incubated individually in the presence of peptides corresponding to each of the five Pap subunit Ntes (PapG lacks an Nte, as it is positioned at the distal end of the pilus and acts only as a donor strand acceptor). The DSE reaction was followed by characterizing the time course of the disappearance of the chaperone–subunit complex by electrospray mass spectrometry. Of the 30 reactions examined, 7 mimicked the interactions that are known to occur in the pilus structure in vivo (referred to as cognate interactions) and the remaining 23 reactions involve non-cognate pairings. The results reveal a striking correlation between the apparent rate of DSE and the nature of the Nte (that is, whether is it cognate or non-cognate for the partner subunit), with the most rapid reactions uniformly occurring with cognate partners. The data suggest that the complementarity between each subunit groove and its cognate Nte plays a substantial part in determining subunit assembly. The range of reactivities between different chaperone and subunit Nte pairs is remarkable, given the structural homology between the subunit folds and the highly conserved Nte binding motif comprising the alternating P1 to P5 hydrophobic residues (Fig. 2f). Further studies identified the residues in the Ntes that are responsible for the specific groove–Nte interaction and found that the P5 residue and the residues immediately adjacent to it have key roles in determining DSE rates30.
A comprehensive survey of the P5 pockets in all Pap subunits (except PapG) was recently published31. PapA and PapK both have well-defined P5 pockets (Figs 2e,3b) and are known to undergo DSE at similar rates. In PapE, the A2–B loop is disordered and so the region around the P5 pocket is thought to be flexible. Remarkably, PapE is known to be a fast-exchanging pilus subunit and undergoes DSE at the highest rate among Pap subunits. By contrast, in PapH and PapF the P5 pocket is obstructed by residues in the A2–B loop (namely, Thr52 in PapH and Pro32 in PapF); PapH is unable to undergo DSE and PapF undergoes DSE at a slow rate. Thus, there seems to be a correlation between the level of accessibility and flexibility of the P5 pocket and the rate of DSE: the more accessible and flexible the region around the P5 pocket, the faster the exchange reaction. The P5 pockets in PapF and PapH were further investigated using molecular dynamics and site-directed mutagenesis. Although in both the PapH and PapF crystal structures the P5 pocket seems to be blocked, molecular dynamics simulations suggest that the P5 pocket of PapF becomes accessible intermittently, whereas that of PapH does not. These results, which were confirmed using site-directed mutagenesis followed by DSE experiments, provide an explanation for why PapF undergoes slow DSE and PapH no DSE at all.
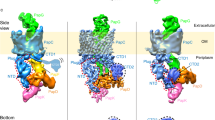
Pilus assembly at the usher. In vivo, subunit polymerization occurs at the outer-membrane usher. The usher recruits binary chaperone–subunit complexes to the outer membrane, mediates their ordered polymerization and is responsible for translocation of the growing pilus to the outer surface4,32,33,34,35. Cryo-EM images of two-dimensional crystals of the UPEC P pilus usher PapC, reconstituted in E. coli lipids, show that PapC forms homodimeric channels. Complementation studies of PapC loss-of-function mutants subsequently established that these dimers constitute the functional unit for the usher36,37. However, although dimer formation is crucial for usher function in vivo, it might not be necessary for usher-mediated fibre formation in vitro38. Outer-membrane ushers are ∼800 residue proteins that comprise four functional domains: an N-terminal, ∼125-residue periplasmic domain, a C-terminal, ∼170-residue periplasmic domain and a large, central, ∼500-residue translocation pore domain that is interrupted by a conserved plug domain of ∼110 residues39,40,41 (Fig. 4a).
a | The domain structure of PapC. The translocation pore and plug domains are shown in blue and magenta, respectively. The amino-terminal domain (NTD) and carboxy-terminal domain (CTD) are also indicated. b | The structure of the FimD N-terminal domain bound to the FimC–FimH complex. c |The structure of the translocation domain of the PapC usher, shown as a ribbon representation, with the barrel in cyan, the single helix in yellow, the trigger hairpin in orange and the plug domain in magenta. d | The cryo-electron microscopy structure of the FimD2–FimH–FimG–FimF–FimC complex.The left panel shows a ribbon diagram of the structure. The two usher protomers are labelled usher 1 and usher 2.Their N-terminal domains are labelled N1 and N2, respectively. In the grey area, a model of FimC bound to FimA and N2 is shown. This model is oriented in such a way that the N-terminal extension (Nte) of FimA is within range for donor strand exchange with the FimF groove (that is, the P5 residue of FimA (not shown) is within interaction distance of the P5 pocket of FimF). The structure of the FimD2–FimH–FimG–FimF–FimC complex is shown in the right panel in a schematic representation. The plug of usher 1 (the secretion pore) is shown in two different conformations: one in which it is still within the lumen of the pore but pushed to the side, and another in which it is pushed out into the periplasm (P). e | The subunit incorporation cycle, mediated by the twinned ushers. An incoming chaperone–subunit (FimC–FimA) complex is recruited by binding to N2 (step 1). Donor strand exchange with FimF causes the FimF-bound chaperone (C1) to be released, and N1 dissociates (steps 2 and 3) to recruit another FimC–FimA complex (step 4), adding it to the N2-bound FimC–FimA complex (step 5). Donor strand exchange then releases N2, which can recruit the next chaperone–subunit complex (step 6). Alternating binding to the released usher N-terminal domains and donor strand exchange with the next chaperone–subunit complex leads to stepwise growth of the pilus (steps 1–6). The plug domains are indicated as P in usher 2 and P′ and P′′ in the alternative orientations shown for usher 1. C, FimC; E, extracellular space; F, FimF; G, FimG; H, FimH; OM, outer membrane. Parts a, d and e are modified, with permission, from Ref. 35 © (2008) Elsevier.
The N-terminal periplasmic domain binds chaperone–subunit complexes with high affinity33,42. The structures of the N-terminal domain of the FimD usher bound to the FimC–FimH chaperone–adhesin complex (Fig. 4b) and to the FimC–FimF chaperone–subunit complex were determined33,43. The main part of the interface between the usher N-terminal domain and both chaperone–subunit complexes is in the chaperone. However, a small part of the interface encompasses subunit-specific interactions, which are likely to explain the differences in affinity for the usher between chaperone–subunit complexes. Indeed, the usher exhibits decreasing affinity according to subunit order in the pilus assembly chain, a process that is believed to be at least in part responsible for subunit ordering4,32,40.
The exact role of the usher C-terminal domain is unclear. Analysis of C-terminal truncation and substitution mutants of the P pilus usher, PapC, demonstrated that the C terminus is required for proper binding of chaperone–subunit complexes to the usher and has an important role in the assembly of complete pili37,38.
The usher translocation pore domain consists of a 24-strand β-pore with an approximate inner diameter of 45 × 25 Å. These dimensions are compatible with the passage of folded pilus subunits35,38 (Fig. 4c). The β-barrel domain is interrupted at strands 6 and 7 by the plug domain, which blocks the channel. The plug domain is required for fibre assembly, as deletion of the plug region leads to the formation of an usher that is unable to assemble pili in vitro38.
Differences in the protease susceptibility of the type 1 pilus usher, FimD, show that the usher undergoes a conformational change on binding the N-terminal domain of the tip adhesin, FimH32,44,45. This adhesin-induced conformational change constitutes a crucial activation step that is required for subunit polymerization and translocation by the usher34,46. Such an activation step must include repositioning of the plug domain, either moving it aside within the translocation pore or rotating it from the pore into the periplasm35. Complementation studies on non-functional papC mutants suggest that C-terminally truncated ushers fail to activate the translocation pore, implicating the C-terminal domain in usher activation events37.
In the absence of the type 1 pilus rod subunit, FimA, pilus assembly stalls in a three-subunit intermediate comprising FimD bound to FimH, and the linker subunits FimG and FimF, with the last incorporated subunit, FimF, still bound to its periplasmic chaperone46. Structural analysis of the FimD–FimH–FimG–FimF–FimC assembly intermediate by cryo-EM reconstruction showed that, remarkably, only a single usher pore in the dimeric usher complex is used for secretion35 (Fig. 4d). FimH extrudes from the pore, FimG resides in the pore and the FimC–FimF complex bound to the N-terminal domain of the usher resides on the periplasmic side, just under the secretion pore. In the isolated complex, the second usher pore lies idle, probably in its non-activated, closed form. The requirement for the second usher to make a functional assembly complex is believed to stem from the need for two chaperone–subunit recruitment sites: in the proposed model, the recruitment of new chaperone–subunit complexes to the assembly platform through their N-terminal domains alternates between the two usher protomers35 (Fig. 4d,e). However, this model is not universally accepted as, at least in vitro, the ushers seem to function as monomers38.
Subunit polymerization and fibre formation at the usher is a process that occurs independently of cellular energy47. In the absence of the usher, subunit polymerization is a spontaneous process that is driven by folding energy stored in the chaperone-bound subunits24,48. Chaperone-bound subunits are maintained in a high-energy folding intermediate that relaxes upon DSE with the Nte of a new incoming subunit24. However, this process is slow and occurs on the scale of hours to days. By contrast, pilus assembly in vivo occurs in a matter of minutes47 and thus it has long been speculated that the usher acts as a polymerization catalyst. A recent study of UPEC type 1 pilus formation directly demonstrated that the usher does indeed increase the rate of subunit polymerization34. In the presence of the FimD usher, in vitro polymerization of FimA is two orders of magnitude faster and occurs in a processive manner, resulting in fewer, longer pili than usher-independent DSE. The observed catalytic function of the usher required pre-incubation with the FimC–FimH chaperone–adhesin complex, confirming the reported FimH-dependent activation step of FimD32,37,45. This step probably involves the gating of the usher secretion pore (discussed above).
As described above, in vitro DSE is a concerted process that starts with the insertion of the Nte of the incoming subunit into the P5 pocket of the subunit that is undergoing strand exchange. Interestingly, the orientation of the last-incorporated chaperone-bound subunit in the cryo-EM reconstruction of the type 1 pilus tip complex is such that its P5 pocket is exposed and points towards the chaperone–subunit recruitment site on the second usher (Fig. 4d). A mechanism of subunit polymerization at the usher was proposed, in which the spontaneous DSE process at the base of the fibre is catalysed by the effect of increased local concentration and the favourable orientation of the Nte and P5 pocket in the attacking and accepting subunits, respectively35 (Fig. 4e).
Adhesin–receptor interaction
Colonization of mucosal surfaces is a key event in bacterial pathogenesis. Bacterial attachment is often the result of the specific recognition of a pilus-associated adhesin by a host receptor; indeed, it has been shown that type 1 pili are essential for UPEC to cause cystitis49,50,51,52. Using scanning and high-resolution EM and a mouse cystitis model, it was demonstrated that adhesive, type 1-piliated UPEC can bind and invade host superficial facet cells. This process is dependent on the FimH adhesin, which is located at the distal end of the pilus and binds mannose residues on host epithelial cells.
The structures of the adhesin domains of FimH from UPEC and enterotoxigenic E. coli (ETEC), PapG from UPEC, and GafD and F17-G in G- and F17-fimbriae from UPEC and ETEC, respectively, show that, despite their lack of sequence similarity, these adhesins have a similar β-barrel jelly-roll fold53,54,55. However, the receptor-binding sites in these adhesins differ markedly and occupy different parts of the structure. The structure of FimH bound to D-mannopyranoside53 revealed that the mannose is buried in a deep, negatively charged pocket at the tip of the receptor-binding domain (Fig. 5a). Residues that are important for the interaction with mannotriose (Man(α1–3)[Man(α1–6)]Man) and D-mannose were confirmed by site-directed mutagenesis. Using these mutants in a mouse model, it was found that the monosaccharide α-D-mannose is the primary bladder cell receptor for type 1-piliated UPEC53,55.
Conversely, when PapGII (one of the three PapG classes, each of which binds different globoseries glycolipids) binds to its receptor, the sugar is bound in a shallow binding pocket formed by three strands and a loop and located on the side of the molecule (Fig. 5b). This interaction is thought to be crucial for the ability of pyelonephritic strains of E. coli to cause kidney disease56. The analogous GafD and F17-G bind the terminal N-acetyl-D-glucosamine (GlcNAc) residues of glycoproteins. The structures of GafD and F17-G in complex with GlcNAc reveal a shallow sugar-binding site, also on the side of the molecule but in a location unrelated to that in PapG. Together, these structures underline the flexibility with which different bacterial strains can establish tropism in infection through the use of a common scaffold for receptor binding.
Conclusions
Gram-negative bacterial pathogens use the CU pathway to assemble pilin subunits into pili. Work on CU pathways over the past decade has used structural biology combined with a multidisciplinary approach to examine the details of protein–protein and protein–carbohydrate interactions in pilus biogenesis and their crucial functions in bacterial pathogenesis. The periplasmic chaperones that are required for pilus assembly were found not only to serve as folding templates for pilus subunits, but also to actively catalyse pilus biogenesis by stabilizing high-energy conformations in the subunits3,19. This work has provided new insights into one of the most basic questions in molecular biology: how do proteins fold into domains that can serve as building blocks for macromolecular assemblies? Current studies are integrating multiple disciplines, ranging from innovative translational research to providing snapshots of molecules caught in the act of triggering disease processes57,58, to unveil unanticipated mechanisms and reshape models of bacterial infections (Box 1). Ultimately, these efforts will lead to a better understanding of infection, hopefully sparking better therapies to treat chronic and recurrent infections (Box 2) and generating models that can be used for the study of an array of human diseases. Work on the CU pathway is generating new insights into some of the most basic principles of protein folding and macromolecular assembly and is providing a paradigm for understanding infectious diseases and developing improved strategies for their treatment and prevention.
References
Fronzes, R., Remaut, H. & Waksman, G. Architectures and biogenesis of non-flagellar protein appendages in Gram-negative bacteria. EMBO J. 27, 2271–2280 (2008).
Sauer, F. G., Remaut, H., Hultgren, S. J. & Waksman, G. Fiber assembly by the chaperone-usher pathway. Biochim. Biophys. Acta 1694, 259–267 (2004).
Barnhart, M. M. et al. PapD-like chaperones provide the missing information for folding of pilin proteins. Proc. Natl Acad. Sci. USA 97, 7709–7714 (2000).
Dodson, K. W., Jacob-Dubuisson, F., Striker, R. T. & Hultgren, S. J. Outer-membrane PapC molecular usher discriminately recognizes periplasmic chaperone-pilus subunit complexes. Proc. Natl Acad. Sci. USA 90, 3670–3674 (1993).
Thanassi, D. G. et al. The PapC usher forms an oligomeric channel: implications for pilus biogenesis across the outer membrane. Proc. Natl Acad. Sci. USA 95, 3146–3151 (1998).
Nuccio, S. P. & Baumler, A. J. Evolution of the chaperone/usher assembly pathway: fimbrial classification goes Greek. Microbiol. Mol. Biol. Rev. 71, 551–575 (2007).
Kuehn, M. J., Heuser, J., Normark, S. & Hultgren, S. J. P pili in uropathogenic E. coli are composite fibres with distinct fibrillar adhesive tips. Nature 356, 252–255 (1992).
Baga, M., Norgren, M. & Normark, S. Biogenesis of E. coli Pap pili: PapH, a minor pilin subunit involved in cell anchoring and length modulation. Cell 49, 241–251 (1987).
Verger, D., Miller, E., Remaut, H., Waksman, G. & Hultgren, S. Molecular mechanism of P pilus termination in uropathogenic Escherichia coli. EMBO Rep. 7, 1228–1232 (2006).
Hahn, E. et al. Exploring the 3D molecular architecture of Escherichia coli type 1 pili. J. Mol. Biol. 323, 845–857 (2002).
Hultgren, S. J. et al. The PapG adhesin of uropathogenic Escherichia coli contains separate regions for receptor binding and for the incorporation into the pilus. Proc. Natl Acad. Sci. USA 86, 4357–4361 (1989).
Jones, C. H., Danese, P. N., Pinkner, J. S., Silhavy, T. J. & Hultgren, S. J. The chaperone-assisted membrane release and folding pathway is sensed by two signal transduction systems. EMBO J. 16, 6394–6406 (1997).
Vetsch, M. et al. Pilus chaperones represent a new type of protein-folding catalyst. Nature 431, 329–333 (2004).
Holmgren, A. & Branden, C. I. Crystal structure of chaperone protein PapD reveals an immunoglobulin fold. Nature 342, 248–251 (1989). This study elucidates the structure of a periplasmic chaperone.
Kuehn, M. J. et al. Structural basis of pilus subunit recognition by the PapD chaperone. Science 262, 1234–1241 (1993). This paper describes the first structure of a complex between a periplasmic chaperone and a peptide derived from a pilin subunit.
Slonim, L. N., Pinkner, J. S., Branden, C. I. & Hultgren, S. J. Interactive surface in the PapD chaperone cleft is conserved in pilus chaperone superfamily and essential in subunit recognition and assembly. EMBO J. 11, 4747–4756 (1992).
Choudhury, D. et al. X-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic Escherichia coli. Science 285, 1061–1066 (1999).
Sauer, F. G. et al. Structural basis of chaperone function and pilus biogenesis. Science 285, 1058–1061 (1999). This article and reference 17 present the structures of chaperone–subunit complexes and formulate the concepts of donor strand complementation for chaperone function and donor strand exchange for subunit assembly. Reference 17 also provides insights into receptor recognition by the type 1 pilus, as it contains the structure of the FimH lectin domain.
Bann, J. G., Pinkner, J. S., Frieden, C. & Hultgren, S. J. Catalysis of protein folding by chaperones in pathogenic bacteria. Proc. Natl Acad. Sci. USA 101, 17389–17393 (2004).
Hung., D. L., Knight, S. D., Woods, R. M., Pinkner, J. S. & Hultgren, S. J. Molecular basis of two subfamilies of immunoglobulin-like chaperones. EMBO J. 15, 3792–3805 (1996).
Zavialov, A. V. et al. Structure and biogenesis of the capsular F1 antigen from Yersinia pestis: preserved folding energy drives fiber formation. Cell 113, 587–596 (2003).
Remaut, H. et al. Donor-strand exchange in chaperone-assisted pilus assembly proceeds through a concerted β strand displacement mechanism. Mol. Cell 22, 831–842 (2006). This paper presents the first evidence that donor strand exchange occurs through a 'zip-in, zip-out' mechanism that is initiated at the P5 pocket.
Anderson, K. L. et al. An atomic resolution model for assembly, architecture, and function of the Dr adhesins. Mol. Cell 15, 647–657 (2004).
Sauer, F. G., Pinkner, J. S., Waksman, G. & Hultgren, S. J. Chaperone priming of pilus subunits facilitates a topological transition that drives fiber formation. Cell 111, 543–551 (2002). This paper and reference 21 reveal the structures of a ternary complex containing a subunit in donor strand complementation with its cognate chaperone and the same subunit in donor strand exchange with a second subunit. Together with reference 17, these studies validate the concept of donor strand exchange.
Vetsch, M. et al. Mechanism of fibre assembly through the chaperone-usher pathway. EMBO Rep. 7, 734–738 (2006).
Lindberg, F., Lund, B., Johansson, L. & Normark, S. Localization of the receptor-binding protein adhesin at the tip of the bacterial pilus. Nature 328, 84–87 (1987).
Jacob-Dubuisson, F., Heuser, J., Dodson, K., Normark, S. & Hultgren, S. Initiation of assembly and association of the structural elements of a bacterial pilus depend on two specialized tip proteins. EMBO J. 12, 837–847 (1993).
Striker, R., Jacob-Dubuisson, F., Freiden, C. & Hultgren, S. J. Stable fiber-forming and nonfiber-forming chaperone-subunit complexes in pilus biogenesis. J. Biol. Chem. 269, 12233–12239 (1994).
Lee, Y. M., Dodson, K. W. & Hultgren, S. J. Adaptor function of PapF depends on donor strand exchange in P-pilus biogenesis of Escherichia coli. J. Bacteriol. 189, 5276–5283 (2007).
Rose, R. J. et al. Unraveling the molecular basis of subunit specificity in P pilus assembly by mass spectrometry. Proc. Natl Acad. Sci. USA 105, 12873–12878 (2008).
Verger, D. et al. Structural determinants of polymerization reactivity of the P pilus adaptor subunit PapF. Structure 16, 1724–1731 (2008).
Saulino, E. T., Thanassi, D. G., Pinkner, J. S. & Hultgren, S. J. Ramifications of kinetic partitioning on usher-mediated pilus biogenesis. EMBO J. 17, 2177–2185 (1998).
Nishiyama, M. et al. Structural basis of chaperone-subunit complex recognition by the type 1 pilus assembly platform FimD. EMBO J. 24, 2075–2086 (2005). This paper describes the first structure of the FimD N-terminal domain bound to a chaperone–subunit complex.
Nishiyama, M., Ishikawa, T., Rechsteiner, H. & Glockshuber, R. Reconstitution of pilus assembly reveals a bacterial outer membrane catalyst. Science 320, 376–379 (2008). This paper describes the first in vitro reconstitution of pilus biogenesis from purified components.
Remaut, H. et al. Fibre formation across the bacterial outer membrane by the chaperone/usher pathway. Cell 133, 640–652 (2008). This paper describes the structure of the translocation usher pore and proposes a general mechanism of usher function on the basis of the pore structure and a cryo-EM structure of the FimD usher bound to a secretion intermediate.
Li, H. et al. The outer membrane usher forms a twin-pore secretion complex. J. Mol. Biol. 344, 1397–1407 (2004).
So, S. S. & Thanassi, D. G. Analysis of the requirements for pilus biogenesis at the outer membrane usher and the function of the usher C-terminus. Mol. Microbiol. 60, 364–375 (2006).
Huang, Y., Smith, B. S., Chen, L. X., Baxter, R. H. & Deisenhofer, J. Insights into pilus assembly and secretion from the structure and functional characterization of usher PapC. Proc. Natl Acad. Sci. USA 106, 7403–7407 (2009).
Thanassi, D. G., Stathopoulos, C., Dodson, K., Geiger, D. & Hultgren, S. J. Bacterial outer membrane ushers contain distinct targeting and assembly domains for pilus biogenesis. J. Bacteriol. 184, 6260–6269 (2002).
Nishiyama, M., Vetsch, M., Puorger, C., Jelesarov, I. & Glockshuber, R. Identification and characterization of the chaperone-subunit complex-binding domain from the type 1 pilus assembly platform FimD. J. Mol. Biol. 330, 513–525 (2003).
Capitani, G., Eidam, O. & Grutter, M. G. Evidence for a novel domain of bacterial outer membrane ushers. Proteins 65, 816–823 (2006).
Ng, T. W., Akman, L., Osisami, M. & Thanassi, D. G. The usher N terminus is the initial targeting site for chaperone-subunit complexes and participates in subsequent pilus biogenesis events. J. Bacteriol. 186, 5321–5331 (2004).
Eidam, O., Dworkowski, F. S., Glockshuber, R., Grutter, M. G. & Capitani, G. Crystal structure of the ternary FimC-FimFt-FimDN complex indicates conserved pilus chaperone-subunit complex recognition by the usher FimD. FEBS Lett. 582, 651–655 (2008).
Munera, D., Palomino, C. & Fernandez, L. A. Specific residues in the N-terminal domain of FimH stimulate type 1 fimbriae assembly in Escherichia coli following the initial binding of the adhesin to FimD usher. Mol. Microbiol. 69, 911–925 (2008).
Munera, D., Hultgren, S. & Fernandez, L. A. Recognition of the N-terminal lectin domain of FimH adhesin by the usher FimD is required for type 1 pilus biogenesis. Mol. Microbiol. 64, 333–346 (2007).
Saulino, E. T., Bullitt, E. & Hultgren, S. J. Snapshots of usher-mediated protein secretion and ordered pilus assembly. Proc. Natl Acad. Sci. USA 97, 9240–9245 (2000).
Jacob-Dubuisson, F., Striker, R. & Hultgren, S. J. Chaperone-assisted self-assembly of pili independent of cellular energy. J. Biol. Chem. 269, 12447–12455 (1994).
Zavialov, A. V. et al. Resolving the energy paradox of chaperone/usher-mediated fibre assembly. Biochem. J. 389, 685–694 (2005).
Mulvey, M. A. et al. Induction and evasion of host defenses by type 1-piliated uropathogenic Escherichia coli. Science 282, 1494–1497 (1998).
Bahrani-Mougeot, F. K. et al. Type 1 fimbriae and extracellular polysaccharides are preeminent uropathogenic Escherichia coli virulence determinants in the murine urinary tract. Mol. Microbiol. 45, 1079–1093 (2002).
Martinez, J. J., Mulvey, M. A., Schilling, J. D., Pinkner, J. S. & Hultgren, S. J. Type 1 pilus-mediated bacterial invasion of bladder epithelial cells. EMBO J. 19, 2803–2812 (2000).
Wu, X. R., Sun, T. T. & Medina, J. J. In vitro binding of type 1-fimbriated Escherichia coli to uroplakins Ia and Ib: relation to urinary tract infections. Proc. Natl Acad. Sci. USA 93, 9630–9635 (1996).
Hung., C. S. et al. Structural basis of tropism of Escherichia coli to the bladder during urinary tract infection. Mol. Microbiol. 44, 903–915 (2002).
Merckel, M. C. et al. The structural basis of receptor-binding by Escherichia coli associated with diarrhea and septicemia. J. Mol. Biol. 331, 897–905 (2003).
Dodson, K. W. et al. Structural basis of the interaction of the pyelonephritic E. coli adhesin to its human kidney receptor. Cell 105, 733–743 (2001).
Roberts, J. A. et al. The Gal(α1-4)Gal-specific tip adhesin of Escherichia coli P-fimbriae is needed for pyelonephritis to occur in the normal urinary tract. Proc. Natl Acad. Sci. USA 91, 11889–11893 (1994).
Rosen, D. A., Hooton, T. M., Stamm, W. E., Humphrey, P. A. & Hultgren, S. J. Detection of intracellular bacterial communities in human urinary tract infection. PLoS Med. 4, e329 (2007).
Henderson, J. P. et al. Quantitative metabolomics reveals an epigenetic blueprint for iron acquisition in uropathogenic Escherichia coli. PLoS Pathog. 5, e1000305 (2009).
Bishop, B. L. et al. Cyclic AMP-regulated exocytosis of Escherichia coli from infected bladder epithelial cells. Nature Med. 13, 625–630 (2007).
Anderson, G. G. et al. Intracellular bacterial biofilm-like pods in urinary tract infections. Science 301, 105–107 (2003).
Mulvey, M. A., Schilling, J. D. & Hultgren, S. J. Establishment of a persistent Escherichia coli reservoir during the acute phase of a bladder infection. Infect. Immun. 69, 4572–4579 (2001).
Justice, S. S. et al. Differentiation and developmental pathways of uropathogenic Escherichia coli in urinary tract pathogenesis. Proc. Natl Acad. Sci. USA 101, 1333–1338 (2004).
Mysorekar, I. U. & Hultgren, S. J. Mechanisms of uropathogenic Escherichia coli persistence and eradication from the urinary tract. Proc. Natl Acad. Sci. USA 103, 14170–14175 (2006).
Schilling, J. D., Mulvey, M. A., Vincent, C. D., Lorenz, R. G. & Hultgren, S. J. Bacterial invasion augments epithelial cytokine responses to Escherichia coli through a lipopolysaccharide-dependent mechanism. J. Immunol. 166, 1148–1155 (2001).
Linder, H., Engberg, I., Baltzer, I. M., Jann, K. & Svanborg-Eden, C. Induction of inflammation by Escherichia coli on the mucosal level: requirement for adherence and endotoxin. Infect. Immun. 56, 1309–1313 (1988).
Hedges, S., Anderson, P., Lidin-Janson, G., de Man, P. & Svanborg, C. Interleukin-6 response to deliberate colonization of the human urinary tract with Gram-negative bacteria. Infect. Immun. 59, 421–427 (1991).
Mysorekar, I. U., Mulvey, M. A., Hultgren, S. J. & Gordon, J. I. Molecular regulation of urothelial renewal and host defenses during infection with uropathogenic Escherichia coli. J. Biol. Chem. 277, 7412–7419 (2002).
Mysorekar, I. U., Isaacson-Schmid, M., Walker, J. N., Mills, J. C. & Hultgren, S. J. Bone morphogenetic protein 4 signaling regulates epithelial renewal in the urinary tract in response to uropathogenic infection. Cell Host Microbe 5, 463–475 (2009).
Langermann, S. et al. Prevention of mucosal Escherichia coli infection by FimH-adhesin-based systemic vaccination. Science 276, 607–611 (1997).
Langermann, S. et al. Vaccination with FimH adhesin protects cynomolgus monkeys from colonization and infection by uropathogenic Escherichia coli. J. Infect. Dis. 181, 774–778 (2000).
Kihlberg, J., Hultgren, S. J., Normark, S. & Magnusson, G. Probing the combining site of the PapG adhesin of uropathogenic Escherichia coli bacteria by synthetic analogues of galabiose. J. Am. Chem. Soc. 111, 6364–6368 (1989).
Ohlsson, J., Jass, J., Uhlin, B. E., Kihlberg, J. & Nilsson, U. J. Discovery of potent inhibitors of PapG adhesins from uropathogenic Escherichia coli through synthesis and evaluation of galabiose derivatives. Chembiochem 3, 772–779 (2002).
Pinkner, J. S. et al. Rationally designed small compounds inhibit pilus biogenesis in uropathogenic bacteria. Proc. Natl Acad. Sci. USA 103, 17897–17902 (2006).
Wellens, A. et al. Intervening with urinary tract infections using anti-adhesives based on the crystal structure of the FimH-oligomannose-3 complex. PLoS ONE 3, e2040 (2008).
Bouckaert, J. et al. Receptor binding studies disclose a novel class of high-affinity inhibitors of the Escherichia coli FimH adhesin. Mol. Microbiol. 55, 441–455 (2005).
Hedenstrom, M. et al. NMR studies of interactions between periplasmic chaperones from uropathogenic E. coli and pilicides that interfere with chaperone function and pilus assembly. Org. Biomol. Chem. 3, 4193–4200 (2005).
Acknowledgements
This work was funded by Medical Research Council grant 85602 to G.W. and US National Institutes of Health grant 49950 to S.J.H.
Author information
Authors and Affiliations
Related links
Related links
DATABASES
Entrez Genome Project
FURTHER INFORMATION
Glossary
- Chaperone–usher pilus
-
A bacterial cell surface appendage that is assembled by the chaperone–usher pathway of pilus biogenesis.
- Curli
-
A type of fimbria that mediates binding to components of the extracellular matrix and is often implicated in biofilm formation.
- Type IV pilus
-
An elongated, flexible appendage that extends from the surface of Gram-negative bacterial cells and is used for adhesion and for cell motility (twitching motility).
- Type III secretion needle
-
A needle-like secretion apparatus in Gram-negative bacteria that forms pores in host membranes and allows the injection of virulence factors from the bacterial cytoplasm into the cytosol of host cells.
- Type IV secretion pilus
-
A pilus that is formed as part of the versatile secretion systems that are found in Gram-negative and Gram-positive bacteria. It can secrete a wide range of substrates, including protein–protein and protein–DNA complexes, and can directly target eukaryotic cells.
- Molecular dynamics
-
A form of computer simulation that calculates the time- dependent behaviour of atoms and molecules, providing information about the motion of the atoms and the resultant conformational changes in the molecules over time or during an interaction.
Rights and permissions
About this article
Cite this article
Waksman, G., Hultgren, S. Structural biology of the chaperone–usher pathway of pilus biogenesis. Nat Rev Microbiol 7, 765–774 (2009). https://doi.org/10.1038/nrmicro2220
Published:
Issue Date:
DOI: https://doi.org/10.1038/nrmicro2220
This article is cited by
-
Association between Moraxella keratitis and advanced glycation end products
Scientific Reports (2024)
-
Predicting the primary infection source of Escherichia coli bacteremia using virulence-associated genes
European Journal of Clinical Microbiology & Infectious Diseases (2024)
-
Deleting chaperone-usher fimbriae operons to improve L-threonine production in Escherichia coli
Systems Microbiology and Biomanufacturing (2024)
-
Probing the oligomeric re-assembling of bacterial fimbriae in vitro: a small-angle X-ray scattering and analytical ultracentrifugation study
European Biophysics Journal (2021)
-
Novel Strategies to Combat Bacterial Biofilms
Molecular Biotechnology (2021)