Key Points
-
Non-flagellar appendages were first observed in Gram-negative bacteria, in the early 1950s. Since then, these structures — known as pili or fimbriae — have been extensively studied. The best characterized are the type I pili, type IV pili and curli pili. All three pilus types are formed by the non-covalent association of pilin subunits into polymeric structures.
-
Pilus-like structures were first detected in the Gram-positive species Corynebacterium renale, in the late 1960s. Subsequently, pili were detected on the surface of other Gram-positive bacteria, including Actinomyces naeslundii, Corynebacterium diphtheriae, Streptococcus parasanguis, Streptococcus salivarius and Streptococcus sanguis. Most recently, in the past year, pili have also been characterized in all three of the principal streptococcal pathogens that cause invasive disease in humans — group A Streptococcus (GAS), group B Streptococcus (GBS) and Streptococcus pneumoniae.
-
A general feature of the Gram-positive pili identified to date is that they comprise three protein subunits, each of which contains an LPXTG amino-acid motif (or a variant of this motif), which is the target of sortase enzymes. Several reports have shown that the pilin subunits are connected by non-disulphide covalent linkages.
-
Pilus assembly in Gram-positives has been most extensively studied in C. diphtheriae. The first step involves the Sec-dependent secretion of the three pilus components, which remain anchored to the cell membrane. The second step is a sortase-dependent reaction in which the cell-anchored proteins are cleaved at the LPXTG motif, forming acyl-enzyme intermediates containing a covalent thioester bond between the sortase enzyme and the pilus subunit. The third and fourth steps involve the oligomerization of the pilus protein subunits and the anchoring of this oligomerized structure to the cell wall. According to this model, pilus growth occurs by the addition of subunits to the base of the pilus. It is worth noting that an alternative model can be envisaged in which pilus growth occurs by the addition of subunits to the top of the growing pilus, although this model would require energy to promote pilus bending, and at present, there are no clues how this energy could be provided.
-
The three pilus components are the backbone or main pilus protein and two ancillary proteins, AP1 and AP2. Two schemes can be envisaged for the incorporation of the ancillary proteins into the pilus structure. The first hypothesis suggests that the ancillary proteins are incorporated into the pilus backbone in the same way as the main subunit is incorporated, with each ancillary protein preceded and followed by the main pilus subunit, and the second hypothesis suggests that ancillary proteins are 'branches' of the pilus backbone.
-
In all Gram-positive species examined so far, the genes encoding the pilus subunits are clustered together in the same genetic locus and might have been acquired by horizontal transfer. The organization of this region in GAS, GBS and S. pneumoniae is discussed.
-
In Gram-negative pathogens, pili are involved in adhesion to host cells, and it is likely that in Gram-positive pathogens pili have a similar role. The authors propose a model for the pilus-mediated adherence of streptococci to cell surfaces.
-
Finally, the authors discuss the potential applications of Gram-positive pili as vaccine candidates.
Abstract
Most bacterial pathogens have long filamentous structures known as pili or fimbriae extending from their surface. These structures are often involved in the initial adhesion of the bacteria to host tissues during colonization. In Gram-negative bacteria, pili are typically formed by non-covalent interactions between pilin subunits. By contrast, the recently discovered pili in Gram-positive pathogens are formed by covalent polymerization of adhesive pilin subunits. Evidence from studies of pili in the three principal streptococcal pathogens of humans indicates that the genes that encode the pilin subunits and the enzymes that are required for the assembly of these subunits into pili have been acquired en bloc by the horizontal transfer of a pathogenicity island.
Similar content being viewed by others
Main
Non-flagellar appendages were first observed in bacteria in the early 1950s, while the outer-membrane surface of Gram-negative pathogens was being scanned with the electron microscope1,2. During the following decade, these filamentous structures were recognized and characterized for several Gram-negative species by two research groups: one led by James Duguid, who referred to these structures as fimbriae2 (derived from the Latin word for fringe); and the other led by Charles Brinton, who preferred to call these structures pili3,4 (derived from the Latin word for hair or fur) (Box 1). To this day, the two terms, fimbriae and pili, are still in use, even though they are synonymous. For the purposes of this Review, we use the term pili for all non-flagellar polymeric cell-surface organelles.
Over the past five decades, several distinct pilus types have been identified, most of which were described and characterized in Gram-negative bacteria. The best characterized of these cell-surface organelles are type I pili5,6 (expressed by enteropathogenic Escherichia coli ), type IV pili7,8 (expressed by E. coli, and Pseudomonas and Neisseria species) and curli pili9,10 (expressed by some strains of E. coli) (Table 1). Under the electron microscope, type I pili appear as peritrichous, rigid, rod-like structures of 1–2 μm in length, and they have a visibly flexible tip that is known to be involved in bacterial interaction with receptors on the host-cell surface11. Type IV pili are a similar length but differ from type I pili in that they appear to be more flexible and often form bundles at polar locations12. Curli pili are, as their name suggests, coiled structures. All three pilus types are formed by the non-covalent association of pilin subunits into regular polymeric structures.
Pilus assembly in Gram-negative bacteria has been well studied and involves the Sec-dependent secretion of the main (backbone) and tip pilin subunits into the periplasmic space, where chaperones prevent them from folding correctly until they reach the outer membrane, which is the site of assembly13. The best-studied pilus types are generated by one of the following mechanisms: the chaperone/usher pathway (type I pili)14,15,16, a type II secretion system (type IV pili)17 or nucleation-dependent polymerization (curli pili)13 (Table 1).
Different types of pilus can have different roles. For example, both type I and type IV pili are involved in adherence to host cells and induction of signalling in these cells18,19. By contrast, only type IV pili allow the transfer of genetic material20, and they are also known to retract, thereby producing a moving force on the bacterium that is known as twitching motility21. A common feature of Gram-negative pili, however, is their role in adhesion to eukaryotic cells. It has been proposed that bacteria use these structures to form an initial association with host cells, which can then be followed by a more 'intimate' attachment that brings the bacterium into proximity to the host-cell surface. Pili are known to adhere to components of the extracellular matrix (ECM)22, as well as to carbohydrate moieties that are present in glycoprotein or glycolipid receptors23,24. Receptor specificity might be important in determining the specificity and tropism of bacteria for particular host cells (Table 1).
Pilus-like structures on the surface of Gram-positive bacteria were first detected in Corynebacterium renale, by electron microscopy25,26. More recently, surface appendages were reported to be present in Actinomyces naeslundii27,28,29 and were subsequently found in other species, including Corynebacterium diphtheriae 30,31, Streptococcus parasanguis (Streptococcus parasanguinis)30, Streptococcus salivarius 32,33 and Streptococcus sanguis (Streptococcus sanguinis)34. Finally, in the past year, pili were also characterized in all three of the principal streptococcal pathogens that cause invasive disease in humans — group A Streptococcus (GAS; that is, Streptococcus pyogenes )35, group B Streptococcus (GBS; that is, Streptococcus agalactiae )36,37 and Streptococcus pneumoniae 38 — in which they have been shown to have key roles in the adhesion and invasion process and in pathogenesis. Interestingly, in at least two of these species, S. agalactiae and S. pyogenes, pilus components seem to be promising candidates for vaccine development35,39. The scope of this Review is to summarize what is known about the structure, assembly and function of pili in Gram-positive bacteria and, in particular, in the three important human streptococcal pathogens, S. pyogenes, S. agalactiae and S. pneumoniae.
Pili in Gram-positive bacteria
In Gram-positive bacteria, two types of pilus-like structure have been identified by electron microscopy. Certain Gram-positive bacteria — for example, Streptococcus gordonii and Streptococcus oralis — are 'decorated' with short, thin rods or fibrils that extend between 70 and 500 nm from the bacterial surface40,41. Much longer (up to 3 μm long) pilus-like structures that appear as flexible rods have been described in the Gram-positive oral pathogens Corynebacterium species and pathogenic streptococci25,26,30,35,36,37,38,42. Early data from studies of oral Gram-positive pathogens indicated that such structures were involved in adhesion and biofilm formation43.
Ton-That and Schneewind were the first to characterize the long rod-like pili in C. diphtheriae30. A general feature of these rod-like pili, as well as the rod-like pili that have been identified in GAS35, GBS36 and S. pneumoniae38, is that they comprise three covalently linked protein subunits, each of which contains an LPXTG amino-acid motif (where X denotes any amino acid) or a variant of this motif, which is the target of sortase enzymes. During pilus formation, specific sortases catalyse the covalent attachment of the pilin subunits to each other and to the peptidoglycan cell wall30. Several reports have shown that the protein components of pilus structures from Gram-positive bacteria are connected by non-disulphide covalent linkages30,44. This structural organization seems to be a peculiar characteristic of Gram-positive bacteria, because covalent bonds have not been detected between the subunits of the pili of Gram-negative bacteria.
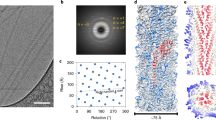
Immunogold electron microscopy using antisera specific for the three pilus components revealed that one protein is the main component (that is, the backbone component) of the structure, and the other two components are ancillary proteins. Antisera specific for the main protein stain the whole length of the pilus structure. Of the two ancillary proteins, antisera specific for ancillary protein 1 (AP1) stain the surface of the bacteria, with occasional staining along the length of the pilus (Fig. 1). In C. diphtheriae and S. pneumoniae, the third and smallest pilus component, AP2, is located along the backbone and at the pilus tip30 (Fig. 1), whereas antisera specific for AP2 from GAS show abundant staining of the bacterial surface and occasional staining that extends from the surface. By contrast, AP2 was not detected in GBS, using immunogold electron microscopy or flow cytometry, although immunoblotting data show that the protein is part of the polymeric structure. These data indicate that AP2 might be hidden inside the GBS pilus structure.
Whole bacterial cells were incubated with polyclonal antibodies conjugated to 5-nm or 10-nm gold particles. a | The image shows the pilus backbone of Streptococcus pn eumoniae stained with gold-labelled antibodies raised against the main pneumococcus pilus component (RrgB). The scale bar represents 100 nm. b | The image shows staining of group B Streptococcus (GBS), with gold-labelled antibodies raised against ancillary protein 1 (GBS104). The scale bar represents 100 nm. c | The image shows immunolabelling of S. pneumoniae pili with antibodies raised against the main pilus protein (RrgB) and ancillary protein 2 (RrgC); 10-nm gold particles are indicated by arrows. The scale bar represents 200 nm.
Assembly of pili in Gram-positive bacteria
Knowledge of the mechanisms of pilus assembly in Gram-positive bacteria initially came from elegant studies of C. diphtheriae by Ton-That and co-workers30,44. The assembly process, which is mediated by sortases catalysing transpeptidase reactions, can be described in four main steps (Fig. 2).
a | In the first step, proteins that contain the amino-acid motif LPXTG (where X denotes any amino acid) are targeted to the cell membrane by Sec-dependent secretion (not shown). This is followed by a sortase-mediated reaction (indicated by the arrows) in which the LPXTG motif is cleaved between the threonine (T) and glycine (G) residues. b | The reaction leads to the formation of an acyl-enzyme intermediate in which a covalent thioester bond is formed between the thiol group of a cysteine residue in the sortase and the carboxyl group of the pilin threonine residue. c | Oligomerization occurs after the nucleophilic attack provided by the ɛ-amino group of the lysine residue in the pilin motif on the cysteine residue of the sortase. d | The thioester bond between the pilin subunit and the sortase is targeted by the amino group of the pentapeptide of lipid II, the precursor of peptidoglycan. e | This leads to the formation of a membrane-associated, covalently linked, elongated pilus. NAG, N-acetyl glucosamine; NAM, N-acetyl muramic acid.
The first step involves the Sec-dependent secretion of the three pilus components (all of which contain an LPXTG motif or a variation of this motif, such as VV/PXTG in the case of the main pilin subunit of GAS, Cpa). Each component remains anchored to the cell membrane, owing to the presence of a membrane-spanning domain at the C terminus. Mutations that alter or abolish this membrane-spanning region prevent these proteins from being retained on the cell surface and promote their secretion44. The second step involves a sortase-dependent reaction in which the cell-anchored proteins are cleaved at the LPXTG motif, between the threonine (T) and glycine (G) residue. This reaction leads to the formation of acyl-enzyme intermediates in which a covalent thioester bond is formed between the thiol group of the cysteine residue located in the catalytic pocket of the sortase and the carboxyl group of the threonine residue in the LPXTG motif of the pilin protein (Fig. 2b). Mutations that result in replacement of this cysteine residue of the sortase or modification of the LPXTG sequence block this enzymatic reaction and impair pilus assembly44. Because sortases are membrane-associated enzymes, the acyl-enzyme derivatives that are formed are retained on the external side of the membrane (Fig. 2b).
The third and fourth steps of the assembly process involve the oligomerization of the pilus protein subunits and the anchoring of the oligomerized structure to the cell wall. These steps require the nucleophilic attack of the thioester bond that links the threonine residue of the pilin subunit to the cysteine residue of the sortase. Although it has not yet been tested experimentally, it is probable that pilus polymerization is achieved while the pilus subunits are attached to the sortase and that the nucleophile is provided by the ɛ-amino group of a specific lysine (K) residue within the pilin motif, WXXXVXVYPKN (where X denotes any amino acid), which has been found in most pilin subunits that have been characterized43. The nucleophilic attack results in cleavage of the thioester bond and concomitant formation of an amide bond between the carbonyl-group carbon of the threonine residue of the pilin subunit (present in the catalytic pocket of the sortase) and the lysine side-chain (ɛ-amino group) of the pilin motif of the neighbouring pilin subunit. This leads to the formation of a membrane-associated covalently linked dimer with a pilin motif that can interact with other sortase-associated pilin subunits, forming an elongated pilus fibre. Ton-That and co-workers have shown that replacing the lysine residue in the pilin motif abolishes the polymerization process, highlighting the importance of this conserved sequence in pilus formation45. According to this model, pilus growth occurs by subunit addition at the base of the pilus (Figs 2,3a), and the length of the pilus depends on the relative abundance of the pilus subunits that are coupled to the membrane-associated sortases. Finally, the association of the membrane-proximal pilus subunit with the cell wall occurs when the thioester bond between the subunit and the sortase is subject to nucleophilic attack by the amino group of the pentapeptide of the peptidoglycan precursor lipid II (Ref. 44), and this leads to the formation of an amide bond between the basal subunit and the cell wall.
a | The first model predicts that ancillary proteins are incorporated into the pilus backbone by the same process as is the main pilus subunit, thereby forming a pilus structure that is interspersed with ancillary-protein-1 or ancillary-protein-2 subunits. b | The second model predicts that ancillary proteins form 'branches' in the pilus shaft and might be more accessible for interaction with host-cell molecules. NAG, N-acetyl glucosamine; NAM, N-acetyl muramic acid.
It should be noted that an alternative model can be envisaged, in which pilus growth occurs by the addition of subunits to the top of the pilus fibre. This model would imply the binding of the first pilus subunit to the peptidoglycan precursor lipid II and the bending of the growing pilus to allow the progressive addition of subunits to the top of the pilus fibre. Although electron-microscopy analysis shows the presence of some pilus structures lying on the bacterial surface, this second model is expected to require energy to promote pilus bending efficiently, and at present, there are no clues about how this energy could be provided to the system.
Several aspects of the pilus-assembly process remain to be elucidated. First, it is still not clear how the different pilus subunits are organized and what determines their order and frequency in the pilus structure. Gene-inactivation experiments in C. diphtheriae have shown that, although the main pilus subunit can polymerize in the absence of the two ancillary proteins, this is not the case for the ancillary proteins30,45, indicating that there are biochemical and/or structural constraints that prevent ancillary proteins from forming homopolymers. As for the mechanism of incorporation of the ancillary proteins into the pilus structure, sortase specificity has a key role. Gram-positive bacteria produce several sortases46, and in most cases, the genomic island that encodes the pilus proteins encodes more than one sortase (Fig. 4). In GBS, the sortases SAG0647 and SAG0648, encoded by pilin island 1, are each specifically required for incorporation of one of the ancillary proteins. Inactivation of the sortase SAG0648 impairs the incorporation of the AP1 GBS104 (also known as SAG0649), and inactivation of the sortase SAG0647 prevents the incorporation of the AP2 GBS52 (also known as SAG0646)37. Interestingly, the main pilus protein, GBS80 (also known as SAG0645), can be polymerized by either of the two sortases, and only inactivation of both sortase genes blocks the reaction completely. In C. diphtheriae strain NCTC13129, only one of the six genome-encoded sortases seems to be sufficient for correct assembly of each pilus type30.
a | Schematic representation of loci that encode group B Streptococcus (GBS; that is, Streptococcus agalactiae) pili. The upper panel shows pilus island 1 (PI-1) in S. agalactiae strain 2603V/R and the same region in S. agalactiae strain 515, which is pilus negative. The operon is flanked by conserved genes sag0633 and sag0652 and direct repeats (DR). In the lower panel, two alleles of PI-2 flanked by conserved genes sag1410 and sag1403 are depicted: PI-2a from S. agalactiae strain 2603V/R, and PI-2b from S. agalactiae strain COH1. b | Genomic organization of the rlrA islet in Streptococcus pneumoniae serotype 4 strain TIGR4 and the laboratory strains R6 and D39, as determined from available sequences. Two insertion sequences (IS 1167) with inverted repeats (IR) flank the locus in pilus-containing strains. (One of the transposases is frame-shifted (fs), whereas an RUP (repeat unit in pneumococcus) element is present in the pilus-negative strain.) The position of the negative-regulator-encoding gene mgrA is indicated. c | Schematic representation of the four fibronectin-binding, collagen-binding, T-antigen (FCT) regions from group A Streptococcus (GAS; that is, S treptococcus pyogenes) strains that belong to seven M types, located between conserved genes spy0123 (which encodes heat-shock protein 33) and spy0136. Dashed black lines represent dimensions that are not to scale.
The specificity of the pilus subunits for the sortases seems to be determined, at least in part, by a third conserved amino-acid sequence, which is located between the LPXTG motif and the pilin motif and is known as the E box, owing to the presence of a highly conserved glutamic-acid residue45. In C. diphtheriae, substitution of the glutamic-acid residue in the E box impairs the incorporation of at least one of the ancillary proteins (SpaB) but does not affect the polymerization of the main pilus protein (SpaA)45. No experimental data have been reported on the role of the E box in other species. However, the E box is conserved in several Gram-positive species and, in particular, is present in the 27 pilin proteins that are encoded by the eight pilus islands that have been characterized so far in GBS (three islands), GAS (four islands) and S. pneumoniae (one island).
The incorporation of the ancillary proteins in the pilus-assembly process is still a mystery, although two schemes can be envisaged (Fig. 3). According to one hypothesis, the ancillary proteins are incorporated into the pilus backbone in the same way as the main subunit is incorporated (Fig. 3a). Because oligomers of ancillary proteins cannot be formed, the only biochemical restriction is that each ancillary protein must be preceded and followed by the main pilus subunit. The other hypothesis predicts that ancillary proteins are 'branches' of the pilus shaft31 (Fig. 3b). The implication of this hypothesis is that all main pilin subunits should carry a fourth conserved amino-acid sequence, which provides the nucleophile to attack the thioester bond that links the ancillary protein subunits to the sortases. Such a conserved sequence must differ from the E box that is involved in sortase specificity not only because the E box does not carry a conserved residue that is likely to attack the acyl derivative but also because, in C. diphtheriae, the substitution of the conserved glutamic-acid residue in SpaA does not affect polymerization of the protein, yet it does affect the incorporation of ancillary proteins45,47.
Pilus islands in the streptococci
In all cases that have been described so far, the genes that encode the pilus proteins are clustered at the same genetic locus (Fig. 4). The genes are transcribed in the same direction, indicating that they are part of an operon. Furthermore, the genes that encode the sortases that are required for pilus assembly are located close to the structural genes and might also be part of the operon.
GBS. There are two separate pilus loci in the GBS genome (Fig. 4a). The first, pilus island 1 (PI-1), is located in a variable part of the genome that consists of ∼16 kb of DNA36 flanked by 11 bp of direct repeats, and this region is found in ∼70% of the GBS strains that have been analysed48. Two conserved genes that are present in all GBS strains that have been analysed flank this DNA region. In strains that lack the region, the flanking genes are contiguous. In addition to the pilus genes, the genomic island contains a gene that encodes an AraC-type transcriptional regulator, as well as a gene (spy0123) that encodes a heat-shock protein (Hsp33) and remnants of transposase-like genes. The overall organization of this genomic region indicates that the complete island was acquired by horizontal DNA transfer.
The second pilus locus in GBS is also located in a variable region of the genome37. There are two variants of this region (PI-2a and PI-2b), which differ in an 11-kb segment of DNA that is flanked by identical conserved genes (sag 1403 and sag 1410). The two variant islands encode distinct pili that have only limited amino-acid-sequence similarity. PI-2a contains, in addition to the genes encoding the three LPXTG-containing proteins and two sortases, a gene that encodes a RogB-type transcriptional regulator. PI-2b lacks the transcriptional regulator but contains a gene that encodes a protein with amino-acid-sequence similarity to the LepA-type signal peptidase of Gram-negative bacteria (Fig. 4a). It is intriguing that in Gram-negative bacteria, this signal peptidase is associated with pilus biosynthesis.
S. pneumoniae. Similarly, in some (but not all) strains of S. pneumoniae, the genes that encode the pilus are contained in a 14-kb pathogenicity island (known as the rlrA islet) (Fig. 4b). This island consists of seven genes, of which rrgA, rrgB and rrgC encode LPXTG-containing proteins. In addition, it contains srtB, srtC and srtD, which encode sortases, and rlrA, which encodes a RofA-like regulator that is a positive regulator of the gene cluster. A transcriptional repressor of the rlrA islet, mgrA, is located outside the islet. The S. pneumoniae pilus island is flanked by IS 1167-containing inverted repeats, which are characteristic of mobile genetic elements, and is absent from the R6 and D39 strains of S. pneumoniae, which are non-pathogenic.
GAS. The pilus operons in GAS also reside on a genomic island. Analysis of the five available GAS genomes allowed the identification of genes that encode surface proteins containing the LPXTG motif and are located in proximity to genes that encode variant sortase enzymes, a common feature of pilus loci (Fig. 4c). The characteristic pilus loci were discovered in a previously described highly variable 11-kb pathogenicity island. This island is known as the fibronectin-binding, collagen-binding, T-antigen (FCT) region, because it contains genes that encode members of a family of ECM-binding proteins, as well as one of the variable antigens that is recognized by antisera of the Lancefield T-serotyping system49. Four classes of FCT region have been described on the basis of gene content and organization. The region is flanked in all cases by two highly conserved genes: spy0123, which encodes the putative chaperone Hsp33; and spy0136 (Fig. 4c). Each of the four FCT regions contains genes that encode distinct pili that react with different T-typing sera. Each region also contains either one or two sortase genes, and three of the regions encode a LepA-type signal peptidase35.
Although the origin and evolution of pilus-like structures in Gram-positive bacteria is still unclear, analysis of the genomic organization of the pilus-encoding operons in invasive streptococci (GAS, GBS and S. pneumoniae) indicates that the capacity to express pili has been acquired by horizontal transfer of a pathogenicity island that carries the genes that are required for the complete biosynthesis of pili. Interestingly, PI-1 of GBS resembles the rlrA islet of S. pneumoniae strain TIGR4 in that it can be present or absent in genomes of different strains. PI-2 of GBS, however, is similar to the GAS island in that it is less complex and contains distinct variable DNA segments. Despite the variability of the islands in the three streptococcal species, there is a similarity in the types of gene that the islands contain. Common features that are encoded by most, but not all, islands include (in addition to the pilus genes) Hsp33, a transcriptional regulator and the LepA-type signal peptidase (Fig. 4).
Although highly variable, the pilus proteins are all clearly related. For example, AP1 from the GBS pilus encoded by PI-1 (GBS104) has 42% identity with AP1 encoded by PI-2a (GBS67; also known as SAG1408) and 50% identity with AP1 of S. pneumoniae (RrgA), whereas its counterpart encoded by GBS PI-2b (SAN1519) has only ∼13% amino-acid-sequence identity with these three proteins. Moreover, the average similarity of pilus variants in the GAS FCT region varies from 23% to 99%. Of note, the main pilus subunit encoded by FCT region 1 (FCT-1) in GAS (T6) has much greater homology to a Streptococcus suis protein (52% identity) than to any GAS protein. Sequence comparisons also show that regions of the N terminus of AP1 proteins encoded by FCT regions are highly similar to the N terminus of the fibronectin-binding proteins encoded in the same genomic regions. This finding indicates that there might be recombination between the two adhesin genes, which could facilitate the generation of variant proteins with different adhesive specificities.
Other Gram-positive organisms. Bioinformatic analysis has provided evidence that genes that encode similar pilus-like proteins (that is, with the previously mentioned motifs and genomic organization) can be found in the genomes of several other Gram-positive bacteria, although experimental data have yet to confirm the presence of these cell-surface structures. Many of these organisms are oral pathogens for which the pili have been characterized (for example, A. naeslundii)28,29, whereas the pili of others (such as Clostridium, Enterococcus, Ruminococcus, Listeria, Staphylococcus, Bacillus, Leuconostoc and Mycobacterium species) have not yet been characterized.
Transcriptional regulators associated with pilus islands. The pilin genes are typically flanked by genes that encode transcriptional regulators, indicative of the existence of 'fine-tuning' mechanisms to alter pilus expression in response to changing environmental conditions. Some of these regulators have been studied in detail, and it has been confirmed that they affect the transcription of pilus genes, as well as the transcription of additional target genes. For example, in GAS, the product of nra in FCT-3 downregulates transcription of cpa (which encodes the collagen-binding pilus component Cpa), as well as the neighbouring gene prtF 2 (which encodes a fibronectin-binding protein)50. In addition, the products of rofA51 and msmR52 are involved in upregulating the transcription of genes that encode GAS pilin components. Furthermore, an msmR homologue (araC) is found near the 5′ boundary of GBS PI-1, and a rofA homologue (rogB) is found upstream of the genes that encode the pilus components in GBS PI-2a. Last, the S. pneumoniae pilus island is flanked by two nra and rofA homologues, rlrA and mgrA. The products of these genes regulate the expression of Rrg pilins, which are known to have a role in colonization of the nasopharynx and lungs by S. pneumoniae38,53,54.
Function of pili and their role in disease
In Gram-negative pathogens, pili have an important role in adhesion and attachment to host cells, an essential step in the pathogenic process. The pili of Gram-positive pathogens are likely to have a similar role. Evidence for this comes from the Gram-positive oral pathogens. For example, A. naeslundii produces two structurally different types of pilus, known as type 1 and type 2. Type 1 pili adhere to the proline-rich salivary proteins that coat the tooth enamel and therefore contribute to the efficiency of oral colonization by A. naeslundi i55. By contrast, type 2 pili have lectin activity and are involved in adhesion to host cells, as well in the formation of biofilms27 that contain A. naeslundii and other oral pathogens, including S. parasanguis56. S. parasanguis also produces pili that mediate adhesion to tooth surfaces coated with proline-rich salivary proteins13.
Pilus components in the invasive streptococci, by contrast, have amino-acid-sequence similarity to members of a large family of proteins known as microbial surface components recognizing adhesive matrix molecules (MSCRAMMs), which are involved in interaction with components of the ECM, including fibronectin, fibrinogen, collagen, laminin and vitronectin57. However, some members of this family are surface-associated proteins that do not assemble into pilus structures. For example, the GAS pilus fibronectin-binding proteins PrtF1 and PrtF2 have 25–28% identity with the GAS pilus collagen-binding protein Cpa. Several members of this family of ECM-binding proteins are directly involved in adhesion to, and invasion of, host cells. The best characterized of these are members of the family of fibronectin-binding proteins that is known to mediate adhesion to host tissues and increase bacterial uptake into non-phagocytic cells58. In addition, Cpa has also been shown to mediate internalization of GAS into HEp-2 cells in an in vitro model of adhesion and invasion59. So, it is likely that — as is the case for Gram-negative pathogens —the pili of Gram-positive bacteria are mainly involved in adhesion and colonization. However, the structure and the biosynthesis of the pili of Gram-negative and Gram-positive bacteria are radically different. It is striking to find such different structures that have probably evolved for similar purposes.
Evidence that the pili of Gram-positive bacteria have a direct role in pathogenesis comes from recent studies of S. pneumoniae38. A strain that carries the S. pneumoniae pilus island was shown to adhere better to lung epithelial cells than did an isogenic strain that lacks the island. The piliated strain also had a competitive advantage over the pilus-negative strain after mixed intranasal challenge and was significantly more virulent in a model of invasive disease. Part of the increase in virulence seemed to result from induction of a stronger inflammatory response. Notably, infection with the piliated strains resulted in significantly higher amounts of tumour-necrosis factor in the bloodstream of mice38. This effect might be a consequence of the higher efficiency of adhesion of piliated bacteria (in contrast to non-piliated bacteria) to cells that are involved in the innate immune response, as well as a consequence of their detection by host-cell pattern-recognition receptors.
The unusual organization of the covalently assembled pili of Gram-positive bacteria could indicate a mechanism of adhesion. In each case, the pili are formed by homopolymerization of the main pilus component, to which a second component (AP1) is added at intervals along the structure and the bacterial surface. In addition, electron-microscopy analysis indicates that the third pilus component in C. diphtheriae might be located at the tip of the pilus. As mentioned earlier, the AP1 components of the GAS pili are collagen-binding proteins and have been shown to function as adhesins50. Given these structures, a plausible model of adhesion can be proposed. In this model, initial contact with the host target cells or ECM is facilitated by the extended nature of the pilus structure, perhaps through an AP2 component at the tip. The adhesive properties of the main pilus and/or AP1 components are such that these components would then be able to interact with host target cells, resulting in the bacteria being drawn closer to host cells by a 'zippering' effect of sequential adhesion down the length of the pilus. This would allow surface-localized AP1 and other non-pilus adhesins on the bacterial surface to interact with their receptors, resulting in intimate attachment to, and colonization of, host tissue (which is shown schematically in Fig. 5). This mechanism is similar to that of the pili of Gram-negative bacteria, several of which have been shown to retract by active disassembly after initial adhesion21, thereby pulling the bacteria closer to the host cells.
a | Free-floating bacteria initiate attachment to host cells by extending their pili towards the apical surface of host cells. This mechanism might involve a tip protein (red). b | Intimate attachment is a secondary process, in which ancillary pilus proteins (green) might be involved in the zipper-like adhesion of pili to host cells, decreasing the distance between the bacterial and host-cell surfaces. c | This intimate attachment leads to colonization of the apical surface of the host cell, a process that is mediated by the expression of high-affinity surface adhesins (blue). In addition, pilus-mediated bacterial aggregation assists the formation of a microbial community in the infected tissue. Colonization as a result of a wide variety of host and pathogen factors, together with increased bacterial-cell density, can lead to an increased innate immune response and inflammation.
In addition to mediating binding to host-cell receptors, pili have been shown to promote the aggregation of other bacteria. Such bacterial co-aggregation might contribute to colonization of tissues and other surfaces, render bacteria more resistant to host defences, and allow beneficial interactions between different bacterial species60,61,62. So, after initial adhesion to, and subsequent intimate interaction with, host cells, the pili could further mediate the co-aggregation of other bacteria during the colonization process (Fig. 5).
Pilus components as vaccine candidates
The components of pili have long been considered as vaccine candidates, owing to their essential role in colonization of the host. In the 1970s, Charles Brinton and colleagues tested purified gonococcal pili as vaccine candidates in humans. Early studies showed that vaccine-induced antibodies could inhibit epithelial attachment of live gonococci and protect volunteers from challenge with bacteria expressing homologous pili63. Unfortunately, this vaccine failed in large-scale efficacy trials64, probably owing to the induction of only a low titre of antibodies that could block colonization and to the high antigenic variation of gonococcus pili in the population. It is interesting to note, however, that the vaccine was administered without an adjuvant, so perhaps the use of a suitable adjuvant would have increased its efficacy.
Brinton and colleagues also experimented with a pilus-based vaccine against enterotoxigenic E. coli (ETEC). In a pig model of maternal immunization, purified pili protected suckling pigs of immunized gilts (immature female pigs) against challenge with virulent ETEC. However, protection was obtained only against strains expressing a homologous pilus protein and not against strains expressing variant pili65.
Encouraging data have recently been obtained from studies in which mice were immunized with the components of the GBS pili, which seem to be more conserved than those of other Gram-positive bacteria. The combination of three pilus components encoded by GBS PI-1 and PI-2a, together with a fourth conserved protein, protected the offspring of immunized mice against lethal challenge with 12 GBS strains, including all nine known serotypes. Interestingly, the level of protection against any particular strain correlated with the level of antigen expression on the bacterial surface. So, one advantage of pilus-based vaccines is that these antigens extend beyond the bacterial cell wall and capsule and are therefore available to interact with protective antibodies. This has been shown for GBS, because the combination vaccine elicited antibodies that were capable of inducing complement-dependent opsono-phagocytic killing by human neutrophils39. That GBS pili seem to be less variable than pili from other bacteria could be because GBS colonizes the gastrointestinal and genital tracts, where there is considerably less immunological pressure than at other sites in the body, and this might contribute to the capacity of a pilus-based vaccine to confer broad coverage.
Components of one of the four described types of GAS pilus have also been shown to confer protection against challenge with a strain expressing the same pilus in a mouse model of infection and disease. Unexpectedly, GAS pili were found to be the antigens recognized by the Lancefield T-serotyping system: that is, the T serotypes. There seem to be only ∼20 T serotypes, probably reflecting strain variability in the pilus proteins. However, the extent of cross-protection between different GAS pilus types is not yet known.
From these data, it is apparent that pilus-based vaccines against Gram-positive pathogens might be more successful than such vaccines directed at their Gram-negative counterparts. It has recently become clear from genomic studies that highly conserved antigens capable of conferring broad coverage might be difficult, if not impossible, to find in most bacterial pathogens39,48. Nevertheless, using combinations of antigens, such as pilus proteins, might achieve broad coverage. Furthermore, combinations of different pilus components might lead to increased immunogenicity and cross-protection, particularly if modern adjuvants are included in the vaccine.
Conclusions
It is becoming increasingly clear that pili are important structures in adhesion and invasion by Gram-positive pathogens, as is the case for Gram-negative pathogens6,10,12,35,56,60. Several questions about the assembly of these unusual covalently linked structures remain to be addressed. Efforts are being made to direct pili assembly in vitro using recombinant pilin subunits and sortases. Together with classical reverse genetics, these studies should lead to the elucidation not only of the assembly process but also of the arrangement of the pilus components in the structures. In vitro adhesion and invasion studies using epithelial-cell monolayers should clarify the role of the pili in these processes and strengthen the rationale for using pilus proteins as vaccine components. Finally, genome sequencing and comparison will lead to a better understanding of the evolution of the pilus-encoding pathogenicity islands and how they have spread through Gram-positive pathogens.
Note added in proof
Since this Review was accepted, a report by Dramsi et al.67 describing pilus assembly in GBS has been published. In this report, AP1 (that is, GBS1478) of the pilus of GBS strain NEM316 was shown to mediate adhesion of the bacteria to human pulmonary epithelial cells.
References
Houwink, A. L. & van Iterson, W. Electron microscopical observations on bacterial cytology; a study on flagellation. Biochim. Biophys. Acta 5, 10–44 (1950).
Duguid, J. P., Smith, I. W., Dempster, G. & Edmunds, P. N. Non-flagellar filamentous appendages (fimbriae) and haemagglutinating activity in Bacterium coli. J. Pathol. Bacteriol. 70, 335–348 (1955). An historic paper showing the presence of fimbriae in E. coli.
Brinton, C. C. Jr. The structure, function, synthesis and genetic control of bacterial pili and a molecular model for DNA and RNA transport in Gram-negative bacteria. Trans. NY Acad. Sci. 27, 1003–1054 (1965). The first description of the function of pili in E. coli.
Brinton, C. C. Jr. Non-flagellar appendages of bacteria. Nature 183, 782–786 (1959).
Duguid, J. P. & Campbell, I. Antigens of the type-1 fimbriae of salmonellae and other enterobacteria. J. Med. Microbiol. 2, 535–553 (1969).
Martinez, J. J., Mulvey, M. A., Schilling, J. D., Pinkner, J. S. & Hultgren, S. J. Type 1 pilus-mediated bacterial invasion of bladder epithelial cells. EMBO J. 19, 2803–2812 (2000).
Koga, T., Ishimoto, K. & Lory, S. Genetic and functional characterization of the gene cluster specifying expression of Pseudomonas aeruginosa pili. Infect. Immun. 61, 1371–1377 (1993).
Freitag, N. E., Seifert, H. S. & Koomey, M. Characterization of the pilF-pilD pilus-assembly locus of Neisseria gonorrhoeae. Mol. Microbiol. 16, 575–586 (1995).
Olsen, A., Jonsson, A. & Normark, S. Fibronectin binding mediated by a novel class of surface organelles on Escherichia coli. Nature 338, 652–655 (1989). Directly showed that adherence to ECM components is mediated by curli pili in E. coli.
Kikuchi, T., Mizunoe, Y., Takade, A., Naito, S. & Yoshida, S. Curli fibers are required for development of biofilm architecture in Escherichia coli K-12 and enhance bacterial adherence to human uroepithelial cells. Microbiol. Immunol. 49, 875–884 (2005).
Hahn, E. et al. Exploring the 3D molecular architecture of Escherichia coli type 1 pili. J. Mol. Biol. 323, 845–857 (2002).
Craig, L., Pique, M. E. & Tainer, J. A. Type IV pilus structure and bacterial pathogenicity. Nature Rev. Microbiol. 2, 363–378 (2004). An excellent review of type IV pili in Gram-negative bacteria.
Wu, H. & Fives-Taylor, P. M. Molecular strategies for fimbrial expression and assembly. Crit. Rev. Oral Biol. Med. 12, 101–115 (2001). An excellent review of pilus-assembly strategies.
Kuehn, M. J. et al. Genetic, biochemical, and structural studies of biogenesis of adhesive pili in bacteria. Methods Enzymol. 236, 282–306 (1994).
Thanassi, D. G., Stathopoulos, C., Dodson, K., Geiger, D. & Hultgren, S. J. Bacterial outer membrane ushers contain distinct targeting and assembly domains for pilus biogenesis. J. Bacteriol. 184, 6260–6269 (2002).
Saulino, E. T., Bullitt, E. & Hultgren, S. J. Snapshots of usher-mediated protein secretion and ordered pilus assembly. Proc. Natl Acad. Sci. USA 97, 9240–9245 (2000).
Strom, M. S. & Lory, S. Structure–function and biogenesis of the type IV pili. Annu. Rev. Microbiol. 47, 565–596 (1993).
Martinez, J. J. & Hultgren, S. J. Requirement of Rho-family GTPases in the invasion of type 1-piliated uropathogenic Escherichia coli. Cell. Microbiol. 4, 19–28 (2002).
Mulvey, M. A. et al. Induction and evasion of host defenses by type 1-piliated uropathogenic Escherichia coli. Science 282, 1494–1497 (1998). Directly showed immune evasion by piliated bacteria.
Chen, I. & Dubnau, D. DNA transport during transformation. Front. Biosci. 8, S544–S556 (2003).
Mattick, J. S. Type IV pili and twitching motility. Annu. Rev. Microbiol. 56, 289–314 (2002).
Amano, A. Molecular interaction of Porphyromonas gingivalis with host cells: implication for the microbial pathogenesis of periodontal disease. J. Periodontol. 74, 90–96 (2003).
Sung, M. A., Fleming, K., Chen, H. A. & Matthews, S. The solution structure of PapGII from uropathogenic Escherichia coli and its recognition of glycolipid receptors. EMBO Rep. 2, 621–627 (2001).
Schweizer, F., Jiao, H., Hindsgaul, O., Wong, W. Y. & Irvin, R. T. Interaction between the pili of Pseudomonas aeruginosa PAK and its carbohydrate receptor β-D-GalNAc(1→4)β-D-Gal analogs. Can. J. Microbiol. 44, 307–311 (1998).
Yanagawa, R., Otsuki, K. & Tokui, T. Electron microscopy of fine structure of Corynebacterium renale with special reference to pili. Jpn J. Vet. Res. 16, 31–37 (1968). The first electron-microscopy study to show the presence of pili in a Corynebacterium species.
Yanagawa, R. & Otsuki, K. Some properties of the pili of Corynebacterium renale. J. Bacteriol. 101, 1063–1069 (1970).
Cisar, J. O. et al. Mutants of Actinomyces viscosus T14V lacking type 1, type 2, or both types of fimbriae. Infect. Immun. 56, 2984–2989 (1988).
Yeung, M. K. & Ragsdale, P. A. Synthesis and function of Actinomyces naeslundii T14V type 1 fimbriae require the expression of additional fimbria-associated genes. Infect. Immun. 65, 2629–2639 (1997).
Yeung, M. K., Donkersloot, J. A., Cisar, J. O. & Ragsdale, P. A. Identification of a gene involved in assembly of Actinomyces naeslundii T14V type 2 fimbriae. Infect. Immun. 66, 1482–1491 (1998).
Ton-That, H. & Schneewind, O. Assembly of pili on the surface of Corynebacterium diphtheriae. Mol. Microbiol. 50, 1429–1438 (2003).
Gaspar, A. H. & Ton-That, H. Assembly of distinct pilus structures on the surface of Corynebacterium diphtheriae. J. Bacteriol. 188, 1526–1533 (2006).
Levesque, C., Vadeboncoeur, C., Chandad, F. & Frenette, M. Streptococcus salivarius fimbriae are composed of a glycoprotein containing a repeated motif assembled into a filamentous nondissociable structure. J. Bacteriol. 183, 2724–2732 (2001).
Weerkamp, A. H., Handley, P. S., Baars, A. & Slot, J. W. Negative staining and immunoelectron microscopy of adhesion-deficient mutants of Streptococcus salivarius reveal that the adhesive protein antigens are separate classes of cell surface fibril. J. Bacteriol. 165, 746–755 (1986).
Fives-Taylor, P. M. & Thompson, D. W. Surface properties of Streptococcus sanguis FW213 mutants nonadherent to saliva-coated hydroxyapatite. Infect. Immun. 47, 752–759 (1985).
Mora, M. et al. Group A Streptococcus produce pilus-like structures containing protective antigens and Lancefield T antigens. Proc. Natl Acad. Sci. USA 102, 15641–15646 (2005).
Lauer, P. et al. Genome analysis reveals pili in Group B Streptococcus. Science 309, 105 (2005).
Rosini, R. et al. Identification of novel genomic islands coding for antigenic pilus-like structures in Streptococcus agalactiae. Mol. Microbiol. 1 June 2006 (doi:10.1111/j.1365-2958.05225.x).
Barocchi, M. A. et al. A pneumococcal pilus influences virulence and host inflammatory responses. Proc. Natl Acad. Sci. USA 103, 2857–2862 (2006).
Maione, D. et al. Identification of a universal group B Streptococcus vaccine by multiple genome screen. Science 309, 148–150 (2005). One of the first reports to provide direct evidence that pili from Gram-positive bacteria are protective antigens.
McNab, R. et al. Cell wall-anchored CshA polypeptide (259 kilodaltons) in Streptococcus gordonii forms surface fibrils that confer hydrophobic and adhesive properties. J. Bacteriol. 181, 3087–3095 (1999).
Willcox, M. D. & Drucker, D. B. Surface structures, co-aggregation and adherence phenomena of Streptococcus oralis and related species. Microbios 59, 19–29 (1989).
Jameson, M. W., Jenkinson, H. F., Parnell, K. & Handley, P. S. Polypeptides associated with tufts of cell-surface fibrils in an oral Streptococcus. Microbiology 141, 2729–2738 (1995).
Yamaguchi, T. & Matsunoshita, N. Isolation and some properties of fimbriae of oral Streptococcus intermedius. Curr. Microbiol. 49, 59–65 (2004).
Ton-That, H. & Schneewind, O. Assembly of pili in Gram-positive bacteria. Trends Microbiol. 12, 228–234 (2004). An excellent review of the pilus-assembly process in Gram-positive bacteria.
Ton-That, H., Marraffini, L. A. & Schneewind, O. Sortases and pilin elements involved in pilus assembly of Corynebacterium diphtheriae. Mol. Microbiol. 53, 251–261 (2004).
Navarre, W. W. & Schneewind, O. Surface proteins of Gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol. Mol. Biol. Rev. 63, 174–229 (1999).
Ton-That, H., Marraffini, L. A. & Schneewind, O. Protein sorting to the cell wall envelope of Gram-positive bacteria. Biochim. Biophys. Acta 1694, 269–278 (2004).
Tettelin, H. et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: implications for the microbial 'pan-genome'. Proc. Natl Acad. Sci. USA 102, 13950–13955 (2005).
Bessen, D. E. & Kalia, A. Genomic localization of a T serotype locus to a recombinatorial zone encoding extracellular matrix-binding proteins in Streptococcus pyogenes. Infect. Immun. 70, 1159–1167 (2002).
Podbielski, A., Woischnik, M., Leonard, B. A. & Schmidt, K. H. Characterization of nra, a global negative regulator gene in group A streptococci. Mol. Microbiol. 31, 1051–1064 (1999).
Kreikemeyer, B., Beckert, S., Braun-Kiewnick, A. & Podbielski, A. Group A streptococcal RofA-type global regulators exhibit a strain-specific genomic presence and regulation pattern. Microbiology 148, 1501–1511 (2002).
Nakata, M., Podbielski, A. & Kreikemeyer, B. MsmR, a specific positive regulator of the Streptococcus pyogenes FCT pathogenicity region and cytolysin-mediated translocation system genes. Mol. Microbiol. 57, 786–803 (2005).
Hava, D. L., Hemsley, C. J. & Camilli, A. Transcriptional regulation in the Streptococcus pneumoniae rlrA pathogenicity islet by RlrA. J. Bacteriol. 185, 413–421 (2003).
Hemsley, C., Joyce, E., Hava, D. L., Kawale, A. & Camilli, A. MgrA, an orthologue of Mga, acts as a transcriptional repressor of the genes within the rlrA pathogenicity islet in Streptococcus pneumoniae. J. Bacteriol. 185, 6640–6647 (2003).
Gibbons, R. J. & Hay, D. I. Human salivary acidic proline-rich proteins and statherin promote the attachment of Actinomyces viscosus LY7 to apatitic surfaces. Infect. Immun. 56, 439–445 (1988).
Whittaker, C. J., Klier, C. M. & Kolenbrander, P. E. Mechanisms of adhesion by oral bacteria. Annu. Rev. Microbiol. 50, 513–552 (1996).
Patti, J. M. & Hook, M. Microbial adhesins recognizing extracellular matrix macromolecules. Curr. Opin. Cell Biol. 6, 752–758 (1994).
Schwarz-Linek, U. et al. Pathogenic bacteria attach to human fibronectin through a tandem β-zipper. Nature 423, 177–181 (2003).
Kreikemeyer, B. et al. Streptococcus pyogenes collagen type I-binding Cpa surface protein. Expression profile, binding characteristics, biological functions, and potential clinical impact. J. Biol. Chem. 280, 33228–33239 (2005).
Kirn, T. J., Lafferty, M. J., Sandoe, C. M. & Taylor, R. K. Delineation of pilin domains required for bacterial association into microcolonies and intestinal colonization by Vibrio cholerae. Mol. Microbiol. 35, 896–910 (2000).
Ochiai, K., Kurita-Ochiai, T., Kamino, Y. & Ikeda, T. Effect of co-aggregation on the pathogenicity of oral bacteria. J. Med. Microbiol. 39, 183–190 (1993).
Paranjpye, R. N. & Strom, M. S. A Vibrio vulnificus type IV pilin contributes to biofilm formation, adherence to epithelial cells, and virulence. Infect. Immun. 73, 1411–1422 (2005).
Brinton, C. C. Jr et al. in Immunobiology of Neisseria gonorrhoeae (eds Brooks G. F., Gotschlich, E. C., Holmes, K. K., Sawyer, W. D. & Young, F. E.) 155–178 (American Society for Microbiology, Washington DC, 1978). One of the first reports to describe the use of pili from Gram-negative bacteria for the development of a vaccine.
Johnson, S. C. et al. Human immunization with Pgh 3-2 gonococcal pilus results in cross-reactive antibody to the cyanogen bromide fragment-2 of pilin. J. Infect. Dis. 163, 128–134 (1991).
Morgan, R. L., Isaacson, R. E., Moon, H. W., Brinton, C. C. & To, C. C. Immunization of suckling pigs against enterotoxigenic Escherichia coli-induced diarrheal disease by vaccinating dams with purified 987 or K99 pili: protection correlates with pilus homology of vaccine and challenge. Infect. Immun. 22, 771–777 (1978).
Dochez, R., Avery, O. T. & Lancefield, R. C. Studies on the biology of Streptococcus. I. Antigenic relationship between strains of Streptococcus haemolyticus. J. Exp. Med. 30, 179–213 (1919).
Dramsi, S. et al. Assembly and role of pili in group B streptococci. Mol. Microbiol. 9 May 2006 (doi:10.1111/j.1365-2958.2006.05190.x).
Acknowledgements
We acknowledge M. Mora, D. Maione and G. Bensi for helpful discussions on the content of the manuscript. A special acknowledgement goes to G. Corsi for art work. We also thank the GAS, GBS and Pneumococcus research groups working on pilus research at Novartis Vaccines & Diagnostics.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors are employed by Novartis Vaccines & Diagnostics.
Related links
Related links
DATABASES
Entrez Genome Project
Glossary
- Sec-dependent secretion
-
A multicomponent system that involves at least seven proteins and mediates the translocation of proteins across biological membranes and into different cellular compartments. Proteins that are secreted through the Sec pathway are produced as precursors that contain a signal sequence and are accompanied to the membrane by a chaperone molecule.
- Chaperone/usher pathway
-
In many Gram-negative bacteria, the biogenesis of P pili, which are encoded by the pap operon, occurs by the chaperone/usher pathway. In this system, PapD binds to, and caps, the pilus-subunit surface, preventing premature aggregation in the periplasm. This chaperone–subunit complex is then targeted to the PapC usher in the outer membrane, which is thought to facilitate uncapping.
- Type II secretion systems
-
Systems that are responsible for the extracellular secretion of toxins and hydrolytic enzymes, many of which contribute to pathogenesis in both plants and animals. Secretion through this type of pathway differs from most other membrane-transport systems in that the substrates of this pathway are folded proteins. This pathway shares many features with the type-IV-pilus biogenesis system, including the ability to assemble a pilus-like structure.
- Nucleation-dependent polymerization
-
A process that is involved in the assembly of curli pili in Escherichia coli. At least five proteins are dedicated to the assembly of these structures on the cell surface. There are two key proteins: CsgA (the major subunit), which can polymerize into curli pili when it comes into contact with CsgB (the minor subunit). It has been proposed that CsgB induces a conformational change in CsgA that nucleates its assembly into pili. Polymerized CsgA could then induce a similar change in the next incoming soluble CsgA and continue the assembly process.
- Extracellular matrix
-
A complex structure that surrounds cells in mammalian tissues. It is composed of three main classes of molecule: structural proteins, such as collagen and elastin; specialized proteins, such as fibronectin and laminin; and proteoglycans. Bacteria use pili and other adhesins to adhere to the extracellular matrix.
- Sortase
-
A transpeptidase that links peptide units on separate chains of peptidoglycan. Specifically, sortases link the threonine (T) residue of the LPXTG motif (where X denotes any amino acid) to the bacterial cell wall by a transpeptidation reaction.
- Lancefield T-serotyping system
-
A method of characterizing group A Streptococcus (GAS) on the basis of serum recognition of a variable trypsin-resistant antigen, the T antigen. T antigens have recently been identified as components of GAS pili.
Rights and permissions
About this article
Cite this article
Telford, J., Barocchi, M., Margarit, I. et al. Pili in Gram-positive pathogens. Nat Rev Microbiol 4, 509–519 (2006). https://doi.org/10.1038/nrmicro1443
Issue Date:
DOI: https://doi.org/10.1038/nrmicro1443
This article is cited by
-
An ester bond underlies the mechanical strength of a pathogen surface protein
Nature Communications (2021)
-
Fully Automated System for Rapid Enrichment and Precise Detection of Enterobacteria Using Magneto-Electrochemical Impedance Measurements
BioChip Journal (2021)
-
Adhesion mechanisms of lactic acid bacteria: conventional and novel approaches for testing
World Journal of Microbiology and Biotechnology (2019)
-
Bent conformation of a backbone pilin N-terminal domain supports a three-stage pilus assembly mechanism
Communications Biology (2018)
-
Insights from the draft genome into the pathogenicity of a clinical isolate of Elizabethkingia meningoseptica Em3
Standards in Genomic Sciences (2017)