Key Points
-
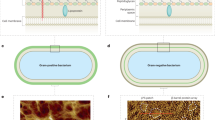
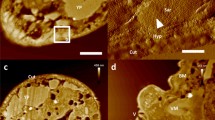
Microbial surfaces form a boundary between microbial cells and their external environment and, as well as this protective function, they actively participate in many cellular interactions. Although electron microscopy techniques have been used to study microbial surfaces to great effect, their use is limited by the fact that they cannot be used to examine specimens in aqueous solution. There was therefore a need to develop new techniques to probe the structure, properties and interactions of microbial surfaces at sub-nanometre resolution; atomic force microscopy (AFM) is one such technique.
-
Unlike conventional microscopy methods, AFM does not use an incident beam to generate an image. Instead, AFM directly measures the physical interaction between the sample and a sensitive probe, or tip. The sample can be moved in three dimensions relative to the tip by a piezoelectric system. The tip is attached to a cantilever, which functions as a small spring and the deflection of the cantilever due to the interaction between the tip and the sample is detected by a laser beam that is focused on the cantilever and reflected onto a photodiode.
-
There are several different operating modes for AFM, including contact mode and tapping mode, which is suitable for imaging 'soft' samples such as bacteria. In force spectroscopy, the forces acting on the instrument tip can be measured to piconewton sensitivity, and this mode can be used to probe the interaction forces between or within biological molecules.
-
There are many different applications for AFM imaging in microbiology research. For example, single membrane proteins can be visualized at sub-nanometre resolution and the surface of living cells can be observed directly as they grow or interact with enzymes. The use of the force spectroscopy mode opens a range of unique possibilities to probe cell-wall elasticity, map cell-surface charges, manipulate individual surface molecules and detect molecular-recognition events.
-
Since its inception in the mid-1980s, the instrumentation and methods of sample preparation for AFM have advanced greatly. The current limitations and technological issues associated with applying AFM to microbiology research are discussed.
Abstract
Our current understanding of microbial surfaces owes much to the development of electron microscopy techniques. Yet, a crucial limitation of electron microscopy is that it cannot be used to examine biological structures directly in aqueous solutions. In recent years, however, atomic force microscopy (AFM) has provided a range of new opportunities for viewing and manipulating microbial surfaces in their native environments. Examples of AFM-based analyses include visualizing conformational changes in single membrane proteins, the real-time observation of cell-surface dynamics, analysing the unfolding of cell-surface proteins and detecting individual cell-surface receptors. These analyses have contributed to our understanding of the structure–function relationships of cell surfaces and will hopefully allow new applications to be developed for AFM in medicine and biotechnology.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Beveridge, T. J. & Graham, L. L. Surface layers of bacteria. Microbiol. Rev. 55, 684–705 (1991).
Sleytr, U. B. & Beveridge, T. J. Bacterial S-layers. Trends Microbiol. 7, 253–260 (1999).
Beveridge, T. J. Structures of Gram-negative cell walls and their derived membrane vesicles. J. Bacteriol. 181, 4725–4733 (1999).
Matias, V. R. F., Al-Amoudi, A., Dubochet, J. & Beveridge, T. J. Cryo-transmission electron microscopy of frozen-hydrated sections of Escherichia coli and Pseudomonas aeruginosa. J. Bacteriol. 185, 6112–6118 (2003).
Mozes, N., Handley, P. S., Busscher, H. J. & Rouxhet, P. G. Microbial Cell Surface Analysis: Structural and Physicochemical Methods. (VCH Publishers, New York, 1991).
Binnig, G., Quate, C. F. & Gerber, C. Atomic force microscope. Phys. Rev. Lett. 56, 930–933 (1986).
Morris, V. J., Kirby, A. R. & Gunning, A. P. Atomic Force Microscopy for Biologists (Imperial College Press, London, 1999).
Jena, B. P. & Hörber, J. K. H. in Methods in Cell Biology Vol. 68, 33–50 (Academic Press, San Diego, 2002).
Butt, H. -J., Jaschke, M. & Ducker, W. Measuring surface forces in aqueous electrolyte solution with the atomic force microscope. Bioelectrochem. Bioenerg. 38, 191–201 (1995).
Heinz, W. F. & Hoh, J. H. Spatially resolved force spectroscopy of biological surfaces using the atomic force microscope. Trends Biotechnol. 17, 143–150 (1999).
Shao, Z., Mou, J., Czajkowsky, D. M., Yang, J. & Yuan, J. -Y. Biological atomic force microscopy: what is achieved and what is needed. Adv. Phys. 45, 1–86 (1996).
Hansma, H. G. & Pietrasanta, L. Atomic force microscopy and other scanning probe microscopies. Curr. Opin. Chem. Biol. 2, 579–584 (1998).
Clausen-Schaumann, H., Seitz, M., Krautbauer, R. & Gaub, H. E. Force spectroscopy with single bio-molecules. Curr. Opin. Chem. Biol. 4, 524–530 (2000).
Engel, A. & Müller, D. J. Observing single biomolecules at work with the atomic force microscope. Nature Struct. Biol. 7, 715–718 (2000).
Fisher, T. E., Marszalek, P. E. & Fernandez, J. M. Stretching single molecules into novel conformations using the atomic force microscope. Nature Struct. Biol. 7, 719–724 (2000).
Hörber, J. K. H. & Miles, M. J. Scanning probe evolution in biology. Science 302, 1002–1005 (2003).
Karrasch, S., Hegerl, R., Hoh, J., Baumeister, W. & Engel, A. Atomic force microscopy produces faithful high-resolution images of protein surfaces in an aqueous environment. Proc. Natl Acad. Sci. USA 91, 836–838 (1994). Early work showing that high-resolution AFM images of native proteins correlate remarkably well with 3-D models determined by established structural methods.
Pum, D. & Sleytr, U. B. Monomolecular reassembly of a crystalline bacterial cell surface layer (S-layer) on untreated and modified silicon surfaces. Supramol. Sci. 2, 193–197 (1995).
Gyorvary, E. S., Stein, O., Pum, D. & Sleytr, U. B. Self-assembly and recrystallization of bacterial S-layer proteins at silicon supports imaged in real time by atomic force microscopy. J. Microsc. 212, 300–306 (2003).
Müller, D. J., Baumeister, W. & Engel, A. Conformational change of the hexagonally packed intermediate layer of Deinococcus radiodurans monitored by atomic force microscopy. J. Bacteriol. 178, 3025–3030 (1996).
Müller, D. J., Schabert, F. A., Büldt, G. & Engel, A. Imaging purple membranes in aqueous solutions at subnanometer resolution by atomic force microscopy. Biophys. J. 68, 1681–1686 (1995).
Schabert, F. A., Henn, C. & Engel, A. Native Escherichia coli OmpF porin surfaces probed by atomic force microscopy. Science 268, 92–94 (1995).
Scheuring, S. et al. High resolution topographs of the Escherichia coli water channel aquaporin Z. EMBO J. 18, 4981–4987 (1999).
Müller, D. J. & Engel, A. Voltage and pH-induced channel closure of porin OmpF visualized by atomic force microscopy. J. Mol. Biol. 285, 1347–1351 (1999). Demonstrates how AFM can be applied to observe conformational changes in single membrane proteins.
Müller, D. J., Fotiadis, D. & Engel, A. Mapping flexible protein domains at subnanometer resolution with the AFM. FEBS Lett. 430, 105–111 (1998).
Ahimou, F., Touhami, A. & Dufrêne, Y. F. Real-time imaging of the surface topography of living yeast cells by atomic force microscopy. Yeast 20, 25–30 (2003).
Dufrêne, Y. F., Boonaert, C. J. P., Gerin, P. A., Asther, M. & Rouxhet, P. G. Direct probing of the surface ultrastructure and molecular interactions of dormant and germinating spores of Phanerochaete chrysosporium. J. Bacteriol. 181, 5350–5354 (1999). The surface of hydrated P. chrysosporium spores was viewed by AFM to a resolution of a few nanometres.
Crawford, S. A., Higgins, M. J., Mulvaney, P. & Wetherbee, R. Nanostructure of the diatom frustule as revealed by atomic force and scanning electron microscopy. J. Phycol. 37, 543–554 (2001).
Schaer-Zammaretti, P. & Ubbink, J. Imaging of lactic acid bacteria with AFM — elasticity and adhesion maps and their relationship to biological and structural data. Ultramicroscopy 97, 199–208 (2003).
Chada, V. G. R., Sanstad, E. A., Wang, R. & Driks, A. Morphogenesis of Bacillus spore surfaces. J. Bacteriol. 185, 6255–6261 (2003).
Doktycz, M. J. et al. AFM imaging of bacteria in liquid media immobilized on gelatin coated mica surfaces. Ultramicroscopy 97, 209–216 (2003).
Touhami, A., Jericho, M. & Beveridge, T. J. Atomic force microscopy of cell growth and division in Staphylococcus aureus. J. Bacteriol. (in the press). A combination of AFM imaging of samples in aqueous solution and thin-section transmission electron microscopy was used to investigate the changes in the bacterial cell wall as the cells grow and divide.
Yao, X. et al. Atomic force microscopy and theoretical considerations of surface properties and turgor pressures of bacteria. Colloids Surf. B Biointerfaces 23, 213–230 (2002).
Kuznetsov, Y. G., Victoria, J. G., Robinson, W. E. & McPherson, A. Atomic force microscopy investigation of human immunodeficiency virus (HIV) and HIV-infected lymphocytes. J. Virol. 77, 11896–11909 (2003).
Malkin, A. J., McPherson, A. & Gershon, P. D. Structure of intracellular mature vaccinia virus visualized by in situ atomic force microscopy. J. Virol. 77, 6332–6340 (2003).
van der Aa, B. C. et al. Stretching cell surface macromolecules by atomic force microscopy. Langmuir 17, 3116–3119 (2001). Shows how force spectroscopy can be applied to living cells to gain insights into the mechanical properties of surface polysaccharides.
Xu, W. et al. Modeling and measuring the elastic properties of an archaeal surface, the sheath of Methanospirillum hungatei, and the implication for methane production. J. Bacteriol. 178, 3106–3112 (1996). A pioneering work that uses AFM to quantitatively measure the elastic properties of a bacterial sheath in relation to its function.
Yao, X., Jericho, M., Pink, D. & Beveridge, T. Thickness and elasticity of Gram-negative murein sacculi measured by atomic force microscopy. J. Bacteriol. 181, 6865–6875 (1999).
Arnoldi, M. et al. Bacterial turgor pressure can be measured by atomic force microscopy. Phys. Rev. E Stat. Phys. Plasmas Fluids Relat. Interdiscip. Topics 62, 1034–1044 (2000).
Touhami, A., Nysten, B. & Dufrêne, Y. F. Nanoscale mapping of the elasticity of microbial cells by atomic force microscopy. Langmuir 19, 4539–4543 (2003).
Ahimou, F., Denis, F. A., Touhami, A. & Dufrêne, Y. F. Probing microbial cell surface charges by atomic force microscopy. Langmuir 18, 9937–9941 (2002).
Dufrêne, Y. F. Direct characterization of the physicochemical properties of fungal spores using functionalized AFM probes. Biophys. J. 78, 3286–3291 (2000).
Fotiadis, D., Scheuring, S., Müller, S. A., Engel, A. & Müller, D. J. Imaging and manipulation of biological structures with the AFM. Micron 33, 385–397 (2002).
Abu-Lail, N. I. & Camesano, T. A. Polysaccharide properties probed with atomic force microscopy. J. Microsc. 212, 217–238 (2003).
Rief, M., Oesterhelt, F., Heymann, B. & Gaub, H. E. Single molecule force spectroscopy on polysaccharides by atomic force microscopy. Science 275, 1295–1297 (1997). Pioneering study demonstrating how the AFM tip can be used in the force spectroscopy mode to pull on single macromolecules, providing new insights into their mechanical and conformational properties.
Marszalek, P. E., Oberhauser, A. F., Pang, Y. P. & Fernandez, J. M. Polysaccharide elasticity governed by chair–boat transitions of the glucopyranose ring. Nature 396, 661–664 (1998).
Abu-Lail, N. I. & Camesano, T. A. Elasticity of Pseudomonas putida KT2442 surface polymers probed with single-molecule force microscopy. Langmuir 18, 4071–4081 (2002). Bacterial cell-surface macromolecules were stretched to learn about their elastic behaviour.
Camesano, T. A. & Abu-Lail, N. I. Heterogeneity in bacterial surface polysaccharides, probed on a single-molecule basis. Biomacromolecules 3, 661–667 (2002).
Müller, D. J., Baumeister, W. & Engel, A. Controlled unzipping of a bacterial surface layer with atomic force microscopy. Proc. Natl Acad. Sci. USA 96, 13170–13174 (1999).
Oesterhelt, F. et al. Unfolding pathways of individual bacteriorhodopsins. Science 288, 143–146 (2000). References 49 and 50 are outstanding papers showing how the combination of AFM imaging and single-molecule force spectroscopy allows researchers to subject membrane proteins to controlled manipulation, thereby providing details of their unfolding pathways and of the forces that anchor them to the membrane.
Scheuring, S. et al. Charting and unzipping the surface layer of Corynebacterium glutamicum with the atomic force microscope. Mol. Microbiol. 44, 675–684 (2002).
Touhami, A., Hoffmann, B., Vasella, A., Denis, F. A. & Dufrêne, Y. F. Aggregation of yeast cells: direct measurement of discrete lectin–carbohydrate interactions. Microbiology 149, 2873–2878 (2003). Biologically modified tips are used to measure the receptor–ligand interaction forces that mediate the aggregation of yeast cells.
Benoit, M., Gabriel, D., Gerisch, G. & Gaub, H. E. Discrete interactions in cell adhesion measured by single-molecule force spectroscopy. Nature Cell Biol. 2, 313–317 (2000). Presents a powerful method for probing cell–cell interactions at the single-molecule level. Uses genetic controls to demonstrate single unbinding events of adhesion proteins.
Pethe, K. et al. Characterization of the heparin-binding site of the mycobacterial heparin-binding hemagglutinin adhesin. J. Biol. Chem. USA 275, 14273–14280 (2000).
Braga, P. C. & Ricci, D. Atomic force microscopy: application to investigation of Escherichia coli morphology before and after exposure to cefodizime. Antimicrob. Agents Chemother. 42, 18–22 (1998).
Kuznetsov, Y. G. et al. Atomic force microscopy investigation of fibroblasts infected with wild-type and mutant murine leukemia virus (MuLV). Biophys. J. 83, 3665–3674 (2002).
Beech, I. B., Smith, J. R., Steele, A. A., Penegar, I. & Campbell, S. A. The use of atomic force microscopy for studying interactions of bacterial biofilms with surfaces. Colloids Surf. B Biointerfaces 23, 231–247 (2002).
Forsythe, J. H., Maurice, P. A. & Hersman, L. E. Attachment of a Pseudomonas sp. to Fe(III)-(hydr)oxide surfaces. Geomicrobiology 15, 293–308 (1998).
Fritz, J. et al. Translating biomolecular recognition into nanomechanics. Science 288, 316–318 (2000).
Müller, D. J., Amrein, M. & Engel, A. Adsorption of biological molecules to a solid support for scanning probe microscopy. J. Struct. Biol. 119, 172–188 (1997).
Gad, M. & Ikai, A. Method for immobilizing microbial cells on gel surface for dynamic AFM studies. Biophys. J. 69, 2226–2233 (1995).
Kasas, S. & Ikai, A. A method for anchoring round shaped cells for atomic force microscope imaging. Biophys. J. 68, 1678–1680 (1995).
Frisbie, C. D., Rozsnyai, L. F., Noy, A., Wrighton, M. S. & Lieber, C. M. Functional group imaging by chemical force microscopy. Science 265, 2071–2074 (1994).
Hinterdorfer, P., Baumgartner, W., Gruber, H. J., Schilcher, K. & Schindler, H. Detection and localization of individual antibody–antigen recognition events by atomic force microscopy. Proc. Natl Acad. Sci. USA 93, 3477–3481 (1996).
Grandbois, M., Dettmann, W., Benoit, M. & Gaub, H. E. Affinity imaging of red blood cells using an atomic force microscope. J. Histochem. Cytochem. 48, 719–724 (2000).
Florin, E. -L., Moy, V. T. & Gaub, H. E. Adhesion forces between individual ligand–receptor pairs. Science 264, 415–417 (1994).
Lee, G. U., Chrisey, L. A. & Colton, R. J. Direct measurement of the forces between complementary strands of DNA. Science 266, 771–773 (1994).
Touhami, A., Hoffmann, B., Vasella, A., Denis, F. & Dufrêne, Y. F. Probing specific lectin–carbohydrate interactions using atomic force microscopy imaging and force measurements. Langmuir 19, 1745–1751 (2003).
Lower, S. K., Hochella, M. F. & Beveridge, T. J. Bacterial recognition of mineral surfaces: nanoscale interactions between Shewanella and α-FeOOH. Science 292, 1360–1363 (2001).
Bowen, W. R., Lovitt, R. W. & Wright, C. J. Atomic force microscopy study of the adhesion of Saccharomyces cerevisiae. J. Coll. Interf. Sci. 237, 54–61 (2001).
Razatos, A., Ong, Y. -L., Sharma, M. M. & Georgiou, G. Molecular determinants of bacterial adhesion monitored by atomic force microscopy. Proc. Natl Acad. Sci. USA 95, 11059–11064 (1998).
Tamayo, J., Humphris, A. D. L., Owen, R. J. & Miles, M. J. High-Q dynamic force microscopy in liquid and its application to living cells. Biophys. J. 81, 526–537 (2001).
Viani, M. B. et al. Probing protein–protein interactions in real time. Nature Struct. Biol. 7, 644–647 (2000).
Ando, T., Kodera, N., Takai, E., Maruyama, D., Saito, K. & Toda, A. A high-speed atomic force microscope for studying biological macromolecules. Proc. Natl Acad. Sci. USA 98, 12468–12472 (2001).
Humphris, A. D. L., Hobbs, J. K. & Miles, M. J. Ultrahigh-speed scanning near-field optical microscopy capable of over 100 frames per second. Appl. Phys. Lett. 83, 6–8 (2003).
Wong, S. S., Joselevich, E., Woolley, A. T., Cheung, C. L. & Lieber, C. M. Covalently functionalized nanotubes as nanometre-sized probes in chemistry and biology. Nature 394, 52–55 (1998).
Acknowledgements
The author is a research associate of the Belgian National Foundation for Scientific Research (FNRS). The support of the FNRS, the Université catholique de Louvain (Special Fund for Research), the Federal Office for Scientific, Technical and Cultural Affairs (Inter-university Poles of Attraction Programme) and the Research Department of Communauté Française de Belgique (Concerted Research Action) is gratefully acknowledged. He thanks nanobiotechnology researchers both in his laboratory and worldwide for sharing exciting experiments and discussions.
Author information
Authors and Affiliations
Ethics declarations
Competing interests
The author declares no competing financial interests.
Related links
Related links
DATABASES
Entrez
SwissProt
FURTHER INFORMATION
Encyclopedia of Life Sciences
Glossary
- CRYO-ELECTRON MICROSCOPY
-
An electron microscopy technique in which the sample is frozen to protect it during imaging.
- ATOMIC FORCE MICROSCOPY
-
(AFM). A relatively new form of microscopy in which a sharp tip is scanned over the surface of a sample, while sensing the interaction force between the tip and the sample. Because AFM does not rely on an incident beam, as in electron or light microscopy, the specimen can be directly observed at high resolution in aqueous solution.
- FORCE SPECTROSCOPY
-
A form of AFM in which the force acting on the tip is measured with piconewton (10−12 N) sensitivity as the tip is pushed towards the sample then retracted from it.
- S-LAYERS
-
Two-dimensional arrays of protein or glycoprotein subunits with a molecular mass between 40,000 and 200,000 Daltons that are common constituents of bacterial cell walls.
- HPI
-
The hexagonally packed intermediate (HPI) layer from Deinococcus radiodurans is an S-layer and was amongst the first S-layer systems to be viewed by AFM at submolecular resolution.
- PORIN
-
A membrane protein that allows the passage of small molecules such as glucose through the membrane.
- CANTILEVER
-
AFM tips are mounted on cantilever beams or triangles, which are typically made of silicon or silicon nitride, that behave like springs. Using Hooke's law, the magnitude of the tip–sample force is proportional to the deflection of the cantilever.
- PIEZOELECTRIC CERAMICS
-
Materials that expand or contract when subjected to a potential difference.
- MICROFABRICATION
-
A range of techniques that are derived from the techniques used in microelectronics to make integrated circuits and which are used to make AFM tips and cantilevers.
- DEFLECTION
-
The vertical bending of the AFM cantilever resulting from the tip–sample interaction force.
- BACTERIORHODOPSIN
-
A light-driven proton pump that is packed into a two-dimensional crystal lattice — known as the purple membrane — and integrated into the plasma membrane of Halobacterium salinarium.
- SEPTAL ANNULUS
-
A structure formed during cell division that corresponds to the growth of wall material into the cytoplasm.
- YOUNG'S MODULUS
-
Young's modulus, or the tensile elastic modulus, is a parameter that reflects the resistance of a material to elongation. The higher the Young's modulus, the larger the force needed to deform the material.
- MUREIN SACCULI
-
Murein sacculus is the term used to refer to the net-like peptidoglycan layer that is found in the cell wall of bacteria.
- BUD SCAR
-
The process by which S. cerevisiae proliferates is known as budding. A ring of chitin is formed between the mother cell and the daughter cell (or bud) and once the bud has been pinched off, a mark is left on the surface of the mother cell that is known as the bud scar. Chitin is the main constituent of the bud scar.
- ISOELECTRIC POINT
-
The isoelectric point (or pI) of a protein is the pH at which the protein has an equal number of positive and negative charges.
- FLOCCULATION
-
A process involving the aggregation of microbial cells. In beer-brewing, yeast flocculation occurs spontaneously near the end of fermentation, thereby providing an easy method to dispose of the cells.
Rights and permissions
About this article
Cite this article
Dufrêne, Y. Using nanotechniques to explore microbial surfaces. Nat Rev Microbiol 2, 451–460 (2004). https://doi.org/10.1038/nrmicro905
Issue Date:
DOI: https://doi.org/10.1038/nrmicro905
This article is cited by
-
Hypertonic stress induced changes of Pseudomonas fluorescens adhesion towards soil minerals studied by AFM
Scientific Reports (2023)
-
A biphasic growth model for cell pole elongation in mycobacteria
Nature Communications (2020)
-
Modified cantilever arrays improve sensitivity and reproducibility of nanomechanical sensing in living cells
Communications Biology (2018)
-
Microfluidic bacterial traps for simultaneous fluorescence and atomic force microscopy
Nano Research (2017)
-
The Immunomodulatory Drug Glatiramer Acetate is Also an Effective Antimicrobial Agent that Kills Gram-negative Bacteria
Scientific Reports (2017)