Abstract
Small molecule drugs are relatively effective in working on ‘drugable’ targets such as GPCRs, ion channels, kinases, proteases, etc but ineffective at blocking protein–protein interactions that represent an emerging class of ‘nondrugable’ central nervous system (CNS) targets. This article provides an overview of novel therapeutic modalities such as biologics (in particular antibodies) and emerging oligonucleotide therapeutics such as antisense, small-interfering RNA, and aptamers. Their key properties, overall strengths and limitations, and their utility as tools for target validation are presented. In addition, issues with regard to CNS targets as it relates to the blood–brain barrier penetration are discussed. Finally, examples of their application as therapeutics for the treatment of pain and some neurological disorders such as Alzheimer's disease, multiple sclerosis, Huntington's disease, and Parkinson's disease are provided.
Similar content being viewed by others
INTRODUCTION
Small molecule drugs have dominated central nervous system (CNS) therapeutic markets for decades. Although small molecules are relatively effective in working at ‘drugable’ targets, such as G-protein-coupled receptors, ion channels, kinases, and proteases, they are ineffective at blocking protein–protein interactions that represent the emerging class of ‘nondrugable’ targets. The implementation of genomic and genetic technologies has led to the identification and validation in cellular and animal models of several promising protein–protein interaction targets for the treatment of neurological and psychiatric disorders. The birth of the biotechnology industry has led to introduction of several new drug categories, including recombinant proteins and therapeutic antibodies in the 1980s and 1990s and the introduction of emerging oligonucleotide therapeutics such as antisense, small-interfering RNA (siRNA), and aptamers over the past decade. Although the relative growth rate of small molecule drugs continues to decline, biologics, and in particular recombinant proteins and monoclonal antibodies (mAbs) have revolutionized the treatment of metabolic and immunological disorders, infectious diseases, and cancer and represent the fastest growing product segment of the pharmaceutical industry over the next decade. In this article, we will provide an overview of some of the tools such as antibodies, aptamers, antisense, and siRNA that are available to address protein interaction targets and discuss their utility for the validation of both drugable and nondrugable targets. Although an in-depth review of these novel therapeutic modalities is beyond the scope of this article, we will provide you with examples of their application for the treatment of pain and some neurological disorders such as Alzheimer's disease (AD), multiple sclerosis (MS), Huntington's disease, and Parkinson's disease to name a few.
TOOLS TO ADDRESS NONDRUGABLE TARGETS
The cellular points of intervention of different therapeutic modalities are shown in Figure 1. A comparison of several of the key properties of antibodies, aptamers, antisense, and siRNA is summarized in Table 1 and detailed in the sections below.
Different therapeutic modalities intervene in different parts of the cell. Antibody (Ab) therapeutics intervene on extracellular proteins, by binding to receptors or ligands to modulate their effects, either by inhibition or stimulation. Aptamers and small molecules bind to extracellular and intracellular proteins to alter their function. Both antagonist and agonist effects are possible on targets such as ion channels, neurotransmitter receptors, and kinases. siRNAs and antisense (ASOs) act further upstream at the mRNA level in the cytoplasm and nucleus, respectively, lowering levels of the corresponding protein.
Antibodies
Since the first therapeutic antibody for a cancer indication was approved by the FDA for marketing in 1997, antibodies have become increasingly important in the fight against cancer, autoimmune, and infectious diseases. To date, 22 unconjugated and 5 conjugated mAbs have been approved for these indications. This number will rapidly escalate as there are over 300 mAbs in preclinical and clinical development with the sale of antibodies expected to reach 48% of all therapeutic protein sales by 2009 (EvaluatePharma, 2008; Filpula, 2007; Wang and He, 2007).
Antibodies provide excellent affinity and specificity of target recognition. Furthermore, because of their relatively large size, in vivo stability, and ability to be sequestered from the blood by interacting with FcRn receptors on endothelial cells, they can have extended pharmacokinetic half-lives (days to weeks) (Daugherty and Mrsny, 2006). The drawbacks for mAbs as therapeutics have included the time and expense of production, tendency to illicit immune responses (ie neutralizing antibodies), inability to access intracellular targets, difficulty in tuning pharmacokinetic characteristics post-production, and formulation issues. The development of new methods and technical innovations has partially alleviated some of these concerns.
Monoclonal antibodies were originally exclusively produced by the hybridoma method in which hybrid cells are produced by the fusion of antibody-producing lymphocytes, acquired from animals after in vivo immunization, with a tumor cell. This method was laborious, costly, and produced mAbs prone to illicit a strong and detrimental immune response against murine mAb sequences from human patients. This issue has been largely overcome by chimerization and humanization of murine antibodies, although even humanized mAbs show at least a low incidence of immunogenicity (Filpula, 2007). Furthermore, using transgenic mice expressing repertoires of human antibody genes has simplified the production of humanized antibodies from mice (Lonberg, 2005). Concurrently, a variety of methods have been developed to discover humanized antibodies by wholly in vitro means. Such methods rely on the screening of large (eg >109) antibody libraries using display technologies with phage, cell-surface, and ribosomal scaffolds (Wark and Hudson, 2006). Although still laborious, display methods potentially increase throughput and provide the ability to tune antibodies subsequent to the initial selection.
Antibodies are composed of distinct modules consisting of different functions. For instance, IgG antibodies are bivalent (Figure 2); containing two proteolytically defined variable Fab fragments that consist of two variable domains (VH and VL) and are capable of binding antigen and one constant Fc fragment that is responsible for recruiting immune cells to targeted tissue and subsequent antibody-dependent cellular cytotoxicity or complement-dependent cytotoxicity. This modular architecture is conducive to recombinant manipulation toward producing therapeutics with altered properties, especially circulating half-lives (Figure 2; Holliger and Hudson, 2005). For example, removal of the Fc module (single-chain antibodies or scFvs) will shorten serum half-life and eliminate potential toxic side effects by inappropriate activation of immune cells. This modality has been clinically validated with pexelizumab, the scFvs anti-C5 compliment inhibitor, in trials against acute myocardial infarction (Carrier et al, 2006). Also, recombinant linking of three or more Fab fragments (triabodies, tetrabodies, etc) can increase avidity and possibly serum half-life. The linked Fab fragments can be directed to distinct molecular targets as was done with an anti-CEA/CD3 bispecific diabody in an effort to stimulate the immune systems attack of colon carcinoma cells (Holliger and Manzke, 1999).
IgG and alternate engineered antibody structures and their effect on circulating half-life (t1/2). Schematic representation of different antibody formats with approximate molecular weights and serum half-lives (β-phase). The classic full-length IgG is shown on the left with various and progressively more radical recombinant versions to the right (From: Holliger and Hudson, Nat Biotechnol 23: 1126–1136, 2005).
Other antibody engineering methods have been developed to deliver drugs/toxins to specific tissues such as has been done with radioisotopes (Huhalov and Chester, 2004) or chemotherapeutics (Lillo et al, 2004). The reverse can also be done to engineer antibodies in a way allowing their delivery to specific sites. One example is to chemically couple an antibody to a pH-sensitive polymer that functions to disrupt membranes at acidic pH. This allows an internalized antibody to dissociate from endosomal vesicles and be released into the cytoplasm (Lackey and Press, 2002). Another particularly elegant example is the construction of a trifunctional fusion antibody as a therapeutic for AD. This tribody contains three modules. The first domain binds the human insulin receptor. The insulin receptor mediates the uptake of insulin by the brain and thus this module allows the entire tribody to enter the brain. The second domain is composed of an scFv antibody that binds the amyloid-β peptide (Aβ). Previous work has shown that anti-Aβ antibodies injected into the brain of AD transgenic mice were capable of inducing the rapid clearance of preexisting amyloid plaques. The final module of the tribody contains the CH2–CH3 interface of the human Fc receptor. IgG molecules are rapidly exported from the brain to blood across the blood–brain barrier via the FcR. With these three binding modules, the Aβ antibody was thus capable of (1) crossing the blood–brain barrier into the brain (influx), (2) disaggregating amyloid plaques by binding of Aβ, and (3) removal of the Aβ by efflux across the blood–brain barrier back to the blood (Boado et al, 2007).
Aptamers
Aptamers are nonnaturally occurring oligonucleotides that bind their targets with high affinity and have the capacity to inhibit protein–protein interactions with potencies similar to what is observed with antibodies (Gold et al, 1995; James, 2000). The general process for the identification and optimization of aptamers is shown in Figure 3. Aptamers are generated by the SELEX (systematic evolution of ligands by exponential enrichment) process where high-affinity ligands are isolated from highly diverse (1013–1015) starting pools by repeated rounds of affinity capture and amplification (Tuerk and Gold, 1990). SELEX can be conducted with nucleic acid pools that are highly stabilized to nucleases relative to DNA and RNA (Lin et al, 1994; Burmeister et al, 2005, 2006). The advantage of using such pools for SELEX is that the resulting aptamers bind their targets with high affinity and may be significantly more stable in biological fluids such as plasma than aptamers selected with DNA or RNA. Each oligonucleotide in the SELEX pool typically contains a 30–40 nucleotide region of randomly generated sequence flanked by 15–20 nucleotide 5′- and 3′- primer binding sites, so aptamer hits emerging from a SELEX experiment are typically 60–80 nucleotides in length. To facilitate efficient manufacture at a scale sufficient for therapeutic development and commercialization, aptamers approximately 40 nucleotides long or shorter are desired. Accordingly, the next step in the discovery process, referred to as ‘minimization’, involves identifying the core element within the hit aptamer sequence that binds the target and retains the desired functionality. Subsequently, aptamers may be optimized for potency and nuclease stability by introducing chemical modifications in the ribose ring, at the phosphodiester linkage or on the base moiety at specific residues (Yang and Gorenstein, 2004; Eaton et al, 1997). Aptamer residence time in vivo may be increased by appending high molecular weight polyethylene glycol (PEG) moieties to the aptamer termini (Watson et al, 2000). By modulating both the composition of the aptamer backbone and the overall molecular weight, the pharmacokinetic properties of an aptamer may be tuned to meet the needs of a particular therapeutic setting (Healy et al, 2004; Wilson and Keefe, 2006). For example, pegaptanib (an anti-vascular endothelial growth factor (VEGF) inhibitor for the treatment of age-related macular degeneration), the first aptamer to be approved for therapeutic use, comprises both a nuclease stable aptamer core and a 40 kDa PEG and is administered once every 6 weeks (Chapman and Beckey, 2006; Ng et al, 2006).
Aptamer discovery process. Schematic representation of the aptamer identification and optimization process. Aptamers are generated by an in vitro selection process referred to as SELEX (systematic evolution of ligands by exponential ). Hit aptamers with the desired in vitro functionality are minimized to a length suitable for chemical manufacture at large scale. Chemical modifications to the minimized hit aptamer are introduced to optimize potency and stability in serum. High molecular weight polyethylene glycol (PEG) is then appended to the aptamer reduce or prevent renal filtration and extend aptamer half-life in vivo.
Aptamer antagonists have been characterized for a wide range of proteins (reviewed in Cload et al, 2006), including cytokines (Rhodes et al, 2000, 2001; Ninichuk et al, 2008), growth factors (Bock et al, 1992; Jellinek et al, 1995; Pagratis et al, 1997; Ruckman et al, 1998; Rusconi et al, 2002), proteases, immunoglobulins (Wiegand et al, 1996), and cell adhesion molecules (Jenison et al, 1998; Watson et al, 2000). SELEX may be conducted against targets presented on the surface of cells (reviewed in Shamah et al, 2008), which allows the opportunity for direct selection of aptamer agonists as well. The first example of an aptamer agonist, targeting the co-stimulatory receptor 4-1BB, with demonstrated efficacy in vivo was recently reported (McNamara et al, 2008).
Although pegaptanib is currently the only approved aptamer, several aptamers are currently in clinical development. For example, phase 1 clinical data for an anti-Factor IXa aptamer (Chan et al, 2008) and an anti-von Willebrand Factor (vWF) aptamer (Gilbert et al, 2007) were recently reported. The anti-vWF aptamer acts through a mechanism unlikely to be achieved by a small molecule, namely blockade of the interaction between the A1 domain of vWF and platelets, a key step in the initiation of platelet aggregation. Given their high affinity and specificity, ability to block protein:protein interactions, aptamers present a promising modality for addressing nondrugable targets.
Antisense
Antisense oligodeoxynucleotides (ASOs) are single-stranded oligodeoxynucleotides of approximately 15–25 nucleotides in length that reduce the synthesis of the target protein. In contrast to antibodies and aptamers that bind to proteins to inhibit their function, ASOs exert their effects further upstream, reducing gene expression at the mRNA level (Figure 1). As a result, ASOs can reduce the gene expression of both extracellular and intracellular targets. After entering the cell and binding to targeted mRNA, ASOs downmodulate gene expression by enabling the RNase-H-mediated degradation of the target mRNA or physically blocking translation of the target mRNA. As chemical modifications hinder RNase-H cleavage, such ASO modifications can be leveraged to favor the physical blockade mechanism. Recently, in a novel and very interesting application of RNA-based strategies, ASOs have been used to alter pre-mRNA processing by targeting motifs that involve in splicing (Lacerra et al, 2000; Lim and Hertel, 2001; Mann et al, 2001; Lu et al, 2005; Madocsai et al, 2005; Alter et al, 2006).
To identify ASOs with high selectivity and potency, careful in vitro selection with multiple cell lines to assess efficacy at the target RNA and/or protein level, and confirmation of mechanism of action and specificity by evaluation of closely related isotypes are critical (Crooke, 2004). Further efficacy and specificity analyses in vivo are then required, along with chemical modifications to promote stability and facilitate delivery. Phosphorothioate modifications have been widely used for achieving increased stability of ASOs in vivo against nucleases present in serum. After intravenous administration, these phosphorothioate-modified ASOs are rapidly distributed to peripheral tissues (distribution half-life of 0.4 h; Cossum et al, 1993), accumulating preferentially in organs such as the liver, kidney, and bone marrow. Other routes of administration such as subcutaneous, intradermal, intravitreal, topical, and inhalation have also been used successfully for in vivo delivery of ASOs to peripheral organs and tissues (Crooke, 2004; Cossum et al, 1994). A consideration with repeat dosing of phosphorothioate-modified ASOs is immune stimulation, as evidenced by splenomegaly and mononuclear cell infiltration in organs such as the liver and kidney (Henry et al, 2000). This is particularly pronounced with oligonucleotides containing a CpG motif that activates the Toll-receptor 9 (TLR9), a member of the Toll-receptor family involved in recognition of pathogen-associated molecular patterns and an innate immune cellular response. Stimulation of TLR9 results in an increased production of proinflammatory cytokines, and activation of adaptive immunity. By avoiding these CpG motifs, the proinflammatory response can be minimized. This immune stimulation can be reduced further with chemical modifications such as 5-methyl cytosine and 2′-methoxyethyl (Henry et al, 2000) but not completely eliminated (Senn et al, 2005). Although delivery formulations such as nanoparticles are being investigated for systemic delivery of ASO to nonneural targets, simple ASO formulations in saline appear to be sufficient for uptake into neurons and activity in vivo. In the nervous system, ASOs have been applied successfully with direct CNS administration in preclinical studies to inhibit multiple molecular targets of interest, including ion channels (Porreca et al, 1999), neurotransmitter receptors (Wahlestedt et al, 1993), growth factors (Porritt et al, 2005), signaling molecules (Sakai et al, 2007), and enzymes (Smith et al, 2006).
Although ASOs represent a useful tool for elucidation of target function in the nervous system, the more than 60 clinical trials of ASOs to date include only one neurological disorder—MS. Vitravene (fomivirsen) was the first antisense drug to receive FDA approval and has been marketed for cytomegalovirus retinitis since 1998. Thus far, the clinical settings in which ASO drug candidates have been tested comprise mostly oncology indications, as well as hypercholesterolemia, diabetes, MS, HIV, HCV, Crohn's disease, ulcerative colitis, rheumatoid arthritis, psoriasis, and asthma. However, an ASO targeting superoxide dismutase 1 (SOD1) has been reported to lower production of mutant SOD1 and prolong survival after intraventricular infusion in a transgenic rat model of ALS (Smith et al, 2006). This ASO, ISIS 333611, is in preclinical development at ISIS and may enter clinical trials for the treatment of ALS in the near future.
siRNA
RNA interference (RNAi) is a naturally occurring cellular process of post-transcriptional gene silencing, discovered in 1998 by Fire and Mello (Fire et al, 1998). Synthetic siRNAs are double-stranded RNAs of 21–25 nucleotides in length that harness this naturally occurring pathway. In contrast to antibodies and aptamers that bind to proteins to inhibit their function, siRNAs (like ASOs) exert their effects further upstream, reducing gene expression at the mRNA level (Figure 1). Consequently, siRNAs offer a powerful solution for inhibiting disease targets from all molecular classes, including those that are difficult to modulate selectively with traditional pharmaceutical approaches employing small molecules or proteins. In RNAi, the target mRNA is enzymatically cleaved, leading to decreased levels of the corresponding protein, with specificity being a key feature of the RNAi mechanism. Synthetic siRNAs leverage the naturally occurring process of RNAi in a consistent and predictable manner with regard to extent and duration of action. In the cytoplasm, siRNAs are processed to single guide (antisense) and passenger (sense) strands (Matranga et al, 2005; Rand et al, 2005), and incorporated into RNA-induced silencing complexes (RISC). The RISC complex seeks out and binds the target mRNA of complementary sequence, resulting in the cleavage of the target mRNA by the enzyme Argonaut 2 in the RISC complex (Hammond et al, 2001). As siRNAs harness an endogenous catalytic mechanism, present in all cells, the potency of siRNAs is extremely high with typical siRNAs exhibiting picomolar EC50's in vitro.
siRNAs have been used successfully for elucidating the in vitro and in vivo function of numerous molecular targets of interest. With careful sequence design and selection, and appropriate chemical modification, siRNAs are not only potent, but also highly specific for the target of interest, stable in vivo, and do not elicit immune responses (Bumcrot et al, 2006). Standard sequence design comprises alignment of mRNAs from rodent, monkey and human, and identification of all possible conserved target regions. If sufficient homology exists across species, then multiple siRNA sequences can be screened in vitro for subsequent testing in preclinical models and potential clinical advancement. These possible target regions are then subjected to a BLAST-like analysis, to select sequences that are unlikely to result in ‘off-targeting’ due to silencing of genes that have partial homology to the gene of interest, especially in critical portions of the target region such as the ‘seed’ region (Jackson et al, 2006). These selected sequences, predicted to be specific for the gene of interest, are then tested empirically in vitro to identify the most potent sequences from dose–response studies in cell culture systems. Another mechanism whereby siRNA duplexes can induce unintended effects is through stimulation of the innate immune system. It has been demonstrated that siRNA duplexes with certain sequence motifs can activate TLR7 (Hornung et al, 2005). However, with the use of appropriate chemical modification, the motifs can be inactivated, eliminating immune stimulation without compromising silencing activity (Judge et al, 2005). These same chemical modifications, 2′-O-methyl substitutions, together with phosphorothioate modifications at the 3′ end, provide the additional benefit of stability against endo- and exonucleases, respectively, that are present in biological fluids (Choung et al, 2006). Thus, with appropriate design, selection, and chemical modification, highly potent and specific siRNAs can be identified for in vivo studies.
As siRNA, antisense, and aptamers are all oligonucleotide based, one can envision combining them to generate bifunctional molecules with enhanced properties. In particular, aptamers that target proteins on the cell surface may be useful as delivery agents for siRNA or antisense oligonucleotides. Recently, an anti-prostate-specific membrane antigen (PSMA) aptamer fused to an anti-PLK1 siRNA was shown to specifically reduce levels of PLK1 in LNCaP cells expressing PSMA on their surfaces (McNamara et al, 2006). In addition, the aptamer-siRNA chimera inhibited tumor growth in a xenograft model. As more aptamers that specifically mediate cell delivery are identified, these aptamer-siRNA (ASO) chimeras may find broad utility.
In vivo studies in the nervous system with siRNAs have included inhibition of well-known molecular targets such as the dopamine and serotonin transporters (Thakker et al, 2004, 2005), P2X3 (Dorn et al, 2004), δ-opioid receptor (Luo et al, 2005), and huntingtin (Wang et al, 2005; DiFiglia et al, 2007). In these studies, only the expected phenotypic effects were reported, supporting the specificity of siRNAs in vivo. Moreover, the use of siRNAs as tools for target validation is practical. For neuronal gene silencing in vivo, siRNA simply formulated in saline and delivered directly to the CNS by intrathecal, intraventricular, or intraparenchymal administration achieves neuronal uptake and inhibition of the gene of interest. Although transfection reagents such as iFECT have been used to enhance local delivery in vivo (Hassani et al, 2005; Luo et al, 2005; Tan et al, 2005; Wang et al, 2005; Kumar et al, 2006; Salahpour et al, 2007), such reagents are not absolutely required for neuronal uptake and have an associated risk of cytotoxicity.
A consideration for clinical translation is that for chronic treatment of neurodegenerative diseases, continuous or repeated long-term infusion of siRNA may be required, which is achievable with implantable infusion devices (see below). Although systemic delivery of siRNA to the CNS would be simpler from a clinical standpoint, the blood–brain barrier represents a formidable hurdle for successful delivery. Recently, Kumar et al (2007) have reported silencing in the CNS after intravenous administration of siRNA complexed to the rabies virus glycoprotein peptide, which was proposed to enable delivery across the blood–brain barrier and neuronal uptake by interaction with the nicotinic acetylcholine receptor. Xia et al (2007) have reported successful delivery to intracranial tumors in rats, using intravenous administration of a biotinylated siRNA attached by a biotin-streptavidin linker to a transferrin receptor antibody. If these results can be optimized for drug development, then a significant step would be taken toward turning siRNAs into CNS therapeutics.
For therapeutic application, not only is the identification of specific and potent siRNAs against any molecular target of interest rapid and efficient, but also synthesis of siRNAs in large quantities is straightforward and siRNAs are highly stable upon storage and during handling. Presently, there are five clinical trials in progress with siRNA candidate therapeutics. An siRNA targeting respiratory syncytial virus (RSV) for the treatment of RSV infection is being tested with local administration by Alnylam Pharmaceuticals. Three clinical trials, by OPKO Health, Allergan/Merck, and Quark/Pfizer, target the wet form of macular degeneration, using intravitreal injection of siRNA against VEGF, VEGF-R1, and RTP801i-14, respectively. The first systemic siRNA clinical trial recently commenced with Quark Pharmaceuticals targeting p53 for the treatment of acute renal failure. Additional clinical programs with siRNA are likely to commence in the near future. Not only do siRNAs represent a powerful tool for inhibiting any molecular target of interest for elucidation of target function, but also the exciting potential of siRNAs for therapeutic intervention is being revealed in ongoing clinical trials.
TARGET VALIDATION
Target validation is the process of determining if a particular protein is directly or indirectly involved in propagating a particular disease and is therefore an appropriate target for new drugs. The most straightforward method for accomplishing this task is to inhibit the protein's function in the context of a relevant disease model. Target validation methods attack a gene's function at every level of expression—DNA, RNA, and protein (Figure 1). Knockout technologies attack gene expression at the DNA level and have been invaluable for the target validation of many proteins. However, constructing gene knockouts in mice can be laborious and the results can be difficult to interpret due to differences between mouse and human biology. Also knockout mice sometimes show a phenotype that is inconsistent with extensive in vitro research (Pich and Epping-Jordan, 1998) possibly due to the action of compensating genes or developmental anomalies.
RNAi and antisense are two target validation approaches that inhibit gene expression at the level of RNA. The strengths and limitations of RNAi, siRNA, and antisense have been described in detail in the preceding sections. Although these techniques which target expression at the DNA or RNA levels are powerful in validating targets of interest, they may have very different effects from drugs that act at the protein level. As proteins often either have more than one function or take part in numerous multiprotein complexes, deletion of the protein through altering mRNA levels can lead to the disruption of numerous pathways. In contrast, antibodies and aptamers target a specific protein–protein interaction.
Validating targets at the protein level is more direct and closely mimics the action of commonly used therapeutics such as small molecule drugs and biologics including antibodies. Antibodies, which bind proteins with high affinity and specificity, are a powerful way to inactivate protein targets and have been used against hundreds of extracellular targets (Lichtlen et al, 2002). However, antibodies are too large to traverse the cell membrane to attack intracellular targets. To circumvent this liability scFv antibodies have been expressed intracellularly (Lo et al, 2008). Intracellular antibodies have been named ‘intrabodies’. Because intrabodies have disulfide bonds that are important for maintaining the proper structure there has been some difficulty in achieving proper folding in the reducing intracellular environment. Auf der Maur et al (2004) ameliorated this problem by using an in vivo screening method to find scFv sequences that are more stable. These and other advances in intrabody technology have allowed the practical use of intrabodies for target validation. For instance, Mclear et al (2008) used intrabodies to suppress neurodegeneration in photoreceptors in a fly model of Huntington's disease and found that the intrabodies were more effective in suppressing this aspect of the disease than Hsp70 overexpression.
Similar to antibodies, nucleic acid aptamers bind to the target at the protein level and have been used extensively for target validation purposes (Pendergrast et al, 2005). Aptamers have many attributes that make them suitable for target validation. They bind with high affinity and also exhibit a degree of specificity that allows them to distinguish between closely related molecules. Like small molecule therapeutics their dosage can be easily adjusted and they exhibit good dose-dependent pharmacokinetic and pharmacodynamic responses. Importantly, an aptamer can inhibit target function in vivo by blocking or knocking out a single domain of a protein while leaving the remainder of the protein functional. Small molecules drugs are also known to have this capability (Aramburu et al, 1999). An example of the use of aptamers for validation of an extracellular target in vivo is a series of experiments using an anti-platelet-derived growth factor (PDGF)-B aptamer (Pietras et al, 2001, 2002). PDGF isoforms are potent mitogens and have been implicated in the abnormally high level of interstitial fluid pressure (IFP) seen in human tumors. This elevated pressure is not conducive to the penetration of drugs into the tumors. PDGF-B inhibition using an aptamer led to decreased IFP levels and concomitantly increased levels of chemotherapeutic agents within the tumor (Pietras et al, 2001). Furthermore, the high-affinity aptamer that was specific to PDGF-B and not PDGF-A was able to decrease IFP in a KAT-4 xenograft model and enhance Taxol uptake and efficacy (Pietras et al, 2002).
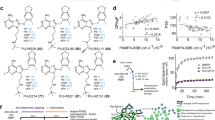
Aptamers, being nucleic acids, are more amenable for use against intracellular targets than antibodies as they can be easily delivered intracellularly by standard transfection techniques or synthesized in vivo with the appropriate expression vectors. Furthermore, they do not need cysteine bonds to fold properly as do protein inhibitors. Some intracellular targets may be refractory to either siRNA and/or aptamer knockout. However, as these methods are both nucleic acid based and act at different levels of expression it was theorized that the two methods could be easily combined and that the combination would be more effective than either method alone. Indeed, Chan et al (2006) knocked down a well-characterized intracellular protein's activity, NFkB, by expressing either aptamers or siRNAs on novel vectors driven by U6 and HIV-1 promoter sequences. The aptamers were encoded into transcripts known to form stable stem and loop structures that localize within the cytoplasm. The best construct placed the aptamer within the natural stable RNA aptamer 7SL with an artificial terminator sequence added on the 3′ end (Figure 4a). This construct diminished NFkB's activity by 64%, about the same as the best siRNA tested (Figure 4b). Interestingly, expression of both aptamers and siRNAs simultaneously suppressed NFkB activity better than either method alone (up to 90%; Figure 4b). The exquisite specificity of aptamers is further exemplified by their ability to discriminate between phosphorylated vs nonphosphorylated kinases (Seiwert et al, 2000) with greater than 10,000-fold specificity suggesting the potential for validating the post-translational active state of a protein toward a particular phenotype.
NFkB activity is most strongly downregulated in vitro in the presence of both siRNA and aptamer inhibitors. (a) Diagram of theoretical transcript generated from aptamer expression construct 7SL-a-p50. (b) Bar graph of results of NFkB-dependent luciferase assay of cells transfected with NFkB-dependent reporter plasmid along with control plasmids for both siRNA and aptamer expressors (siRNA CON+7SL), p50-specific siRNA, and aptamer control plasmid (siRNA2+7SL), p50-specific aptamer plus siRNA control plasmid (siRNA CON+7SL-a-p50), and plasmids expressing both p50-specific siRNA and aptamer (siRNA 2+7SL-a-p50). (From: Pendergrast et al, J BioM Tech 16:224–234, 2005)
ADDRESSING THE BLOOD–BRAIN BARRIER
With few exceptions, small molecules have been the mainstay for the treatment of psychiatric and neurological disorders as well as in pain management. In addition to the obvious benefits of oral administration that are ideal for treatment of chronic CNS conditions, small molecules (with appropriate characteristics) have the ability to cross the blood–brain barrier to access the brain and/or spinal cord. On the other hand, small molecules are limited by their ability to disrupt protein–protein interactions, which limit their utility to the drugable classes of targets that have been reviewed elsewhere in this volume.
The novel therapeutic modalities such as antibodies and oligonucleotide therapeutics such as aptamers, antisense, and siRNA have the distinct advantage of disrupting protein–protein interactions but have some limitations with regard to their ADME (absorption, distribution, metabolism, and excretion) properties that are especially relevant to the treatment of CNS disorders. First, all of these therapeutic modalities are not ideal for oral delivery due to poor absorption from the gastrointestinal tract and are thus limited to intrathecal, intraventricular, intraparenchymal, intravenous, subcutaneous, or intramuscular routes of administration to access the blood or tissue compartments (see Table 1). The requirement for more ‘invasive’ routes of administration is somewhat compensated by the relatively longer half-lives of antibodies relative to small molecules. The half-life of antibodies can range from days to weeks requiring relative infrequent dosing. Breakthroughs in chemical modifications of oligonucleotide therapeutics to make them resistant to serum and tissue nuclease degradation have increased their half-lives in circulation and tissues to several hours to days. Furthermore, through the conjugation of the oligonucleotides to high molecular weight PEGs (see Aptamers above), renal elimination of aptamers has been reduced resulting in half-lives of several days that makes it feasible for potentially once a week administration (Healy et al, 2004). Moreover, RNAi effects have been reported in peripheral organs to persist for up to 4 weeks following termination of siRNA administration (Bartlett and Davis, 2007). The biggest challenge for antibody and oligonucleotide therapeutics is their relative inability to effectively cross the blood–brain barrier. Figure 5 shows the biodistribution of an intravenously administered radiolabeled aptamer (without PEG conjugation) in rat. As can be seen from the autoradiographic image, the aptamer distributes readily to highly perfused organs such as the kidneys, liver, spleen, heart and lungs, and to the lymph nodes (Cload et al, 2006). On the other hand, none of the aptamers showed a propensity to traverse the blood–brain barrier. Similar biodistribution patterns have also been reported for antibodies; although some studies report some limited access to the brain (see Alzheimer's Disease section below). Thus, the utility of these novel therapeutic modalities for the effective treatment of CNS disorders is currently limited to access of relevant targets outside the blood–brain barrier (eg Aβ neutralization approaches in the periphery or limited access to the brain; pain targets in the periphery), addressing diseases with a compromised blood–brain barrier such as MS or direct delivery of the therapeutic to brain using implantable delivery devices.
Aptamer distribution. Quantitative whole-body autoradiograph images of a CD-1 mouse 3 h after single intravenous bolus administration of an unconjugated 3H-labeled aptamer (Cload et al, 2006).
The feasibility of delivering proteins across the blood–brain barrier has been demonstrated using antibody and endocytotic pathways targeting transferrin receptors and low-density lipoprotein receptors that are highly concentrated on brain blood vessels. Carrier systems involving mAbs directed against the transferrin receptor have been utilized to transport large proteins such as nerve growth factor (NGF) across the blood–brain barrier (Granholm et al, 1994). More recently, Spencer and Verma (2007) reported the use of the lentivirus vector system to deliver the lysosomal enzyme glucocerebrosidase and a secreted form of GFP to the neurons and astrocytes in the CNS using a fusion of the low-density lipoprotein receptor-binding domain of the apolipoprotein B to the targeted protein.
Implantable infusion systems for direct delivery of therapeutics to the CNS are being used for intrathecal administration of pain medications and have also been used in clinical trials for intraputamenal administration of a therapeutic candidate for Parkinson's disease. The Medtronic SynchroMed infusion system is an implantable, programmable drug infusion system consisting of a pump, catheter, and programmer that is approved for intrathecal delivery of therapeutics such as Prialt and morphine for the treatment of chronic, intractable pain. This infusion system was also used recently in clinical trials of glial cell line-derived neurotrophic factor (GDNF) for the treatment of Parkinson's disease (Patel et al, 2005; Lang et al, 2006; Slevin et al, 2006) where GDNF was continuously infused for 6 months to 2 years. Although differing efficacy results were obtained in these studies, the trials clearly provide a precedent for the use of implantable infusion systems for direct intraparenchymal delivery of therapeutics to the CNS. Such infusion systems will be valuable for direct CNS delivery of novel therapeutics such as proteins, antibodies, and oligonucleotides. Alnylam Pharmaceuticals and Medtronic are currently co-developing a therapeutic for the treatment of Huntington's disease that is a drug–device combination consisting of an siRNA targeting huntingtin and an implantable infusion system.
EXAMPLES OF CNS DISORDERS ADDRESSED BY NOVEL THERAPEUTIC MODALITIES
Alzheimer's Disease: β-Amyloid Neutralization Approaches
AD, the major cause of dementia in the older population, affects approximately 5.1 million people in the United States (Alzheimer's Disease Facts and Figures 2007, Alzheimer's Association, www.alz.org). Approved drugs for AD, such as the acetylcholinesterase inhibitors Aricept (clonepezil) and Exelon (rivastigmine), alleviate symptoms of the disease such as memory loss but none currently on the market impacts the underlying disease progression. AD is characterized by the presence neuritic plaques composed primarily of the 42 and 40 amino-acid Aβ peptide, products of protease cleavage of the amyloid precursor protein (APP) by the γ- and β-secretases (Selkoe, 2001; Selkoe and Schenk, 2003).
Animal models (murine) such as the PDAPP mouse that expresses an APP mutant (V717F), which results in an increase in the production of Aβ(1−42), recapitulate AD-like plaque pathology, and the concomitant behavioral aspects of the disease (Games et al, 1995). These mouse models have been used extensively to study potential anti-Aβ therapies for AD. In an early study, Schenk et al (1999) demonstrated that immunization of young PDAPP mice with Aβ(1−42) peptide resulted in production of anti-Aβ antibodies and prevented the development of Aβ plaques, dystrophic neurites, and astrocytosis that are characteristic of untreated animals and AD patients. Importantly, immunization of older animals with established plaques yielded similar results.
Several groups have studied the impact of passive immunization of AD mice with anti-Aβ antibodies (Figure 6). Bard et al (2000) have raised anti-Aβ mAbs using peptides representing the N-terminal through the C-terminal residues of Aβ(1−42) as immunogens and evaluated their effects on pathology in a mouse model of AD. Peripheral administration of antibodies to the N terminus of Aβ(1−42) for 6 months dramatically reduced plaque burden in PDAPP mice. Cryostat sections of the brain stained positively with fluorescently labeled anti-mouse antibodies, suggesting that the anti-Aβ antibodies were able to enter the brain. In an ex vivo assay, primary microglial cells were cultured with sections of brain from PDAPP or human AD patients' brains in the presence of various anti-Aβ antibodies. Antibodies that had caused plaque reduction in vivo also induced plaque phagocytosis. Further characterization suggested that the ability of the antibody to bind to Aβ plaques rather than soluble Aβ correlated with induction of plaque phagocytosis ex vivo and reduction of plaque burden in vivo. A F(ab')2 fragment of one efficacious antibody, 3D6, was able to bind plaque but did not induce phagocytosis, suggesting an Fc-receptor-mediated mechanism. In contrast, Bacskai et al (2002) demonstrated that the F(ab')2 fragment of 3D6 was as effective as the full-length antibody at removing plaque deposits from the brains of Tg2576 APP transgenic mice (Holcomb et al, 1998) after topical administration to the surface of the brain. The behavioral impact of Aβ plaque reduction was assessed in PDAPP mice treated with peripherally administered 10D5, a plaque-clearing antibody (Bard et al, 2000); these animals showed improved spatial learning and reduction of Aβ fibrils in the hippocampus (Hartman et al, 2005).
Amyloid-β immunotherapy approach. Anti-Aβ antibodies may prevent or reverse Aβ accumulation in the brain. (http://www.elan.com/Images/FINAL%20Slide%20%231%20MOA_tcm3-6288.pdf).
Antibodies raised against the C terminus of Aβ(1−42) have also demonstrated efficacy in AD transgenic mouse models. Wilcock et al (2004a) administered an anti-Aβ antibody RN2286 (raised against Aβ28−40) for 1, 2, or 3 months to older Tg2576 APP mice to study the impact of treatment on animals with existing plaques. Antibody binding to plaque was detected, and treatment with RN2286 resulted in a reduction in plaque load, microglial activation at early time points, and an increase in circulating Aβ levels (Wilcock et al, 2004a). In a separate study, weekly peripheral administration of RN2286 to Tg2576 APP mice resulted in complete reversal of learning and memory deficits observed in untreated Tg2576 mice as measured by radial-arm water-maze testing (Wilcock et al, 2004b). The requirement for intact antibody effector function to the plaque-clearing action of anti-C-terminal antibodies has been investigated. The impact of treatment with native and deglycosylated forms of 2H6, an antibody to the C terminus of Aβ, has been studied in the Tg2576 mouse AD model (Carty et al, 2006). Intracranial administration with native 2H6 resulted in activation of microglia and upregulation of FcγRII and FcγRIII whereas similar treatment with the deglycosylated antibody, which does not bind with high affinity to the Fc receptors, did not result in microglial activation or upregulation of the receptors. In both cases, treatment resulted in clearance of compact and diffuse plaques from the hippocampus and frontal cortex, which suggests that although phagocytosis may be involved in plaque clearance, it may not be required.
The results described above suggest that treatment with anti-Aβ antibodies may lower plaque burden through an active mechanism within the brain involving microglial-mediated phagocytosis or perhaps disintegration of amyloid plaques initiated by antibody binding. A second series of papers suggest that agents that bind and sequester soluble Aβ may show promise as well, although acting through a different mechanism. DeMattos et al (2001) showed that peripheral administration of m266, an anti-Aβ antibody against amino acids 13–28 of Aβ (Seubert et al, 1992) to PDAPP mice between the ages of 4 and 9 months reduced plaque burden in the hippocampus. In addition, m266 resulted in a 1000-fold increase in the plasma concentration of Aβ within 24 h after administration. m266 binds with high affinity to soluble (nonfibrillar) Aβ peptide and no binding of m266 to plaques was observed. Furthermore, the measured flux of the m266 antibody into the CNS was not sufficient to account for the increase in plasma Aβ. The authors propose that m266 shifts the equilibrium of Aβ between the CNS and the periphery, causing the accumulation of Aβ in the plasma. In a separate study, 24-month-old PDAPP mice treated with m266 for 6 weeks showed memory deficit reversals in object recognition and hole board learning and memory tasks, although their Aβ burden in the brain was not decreased (Dodart et al, 2002). Interestingly, acute treatment of 11-month-old PDAPP mice with m266 resulted in a dose-dependent improvement in object recognition that correlated with an increase of soluble Aβ in the plasma. Building upon that intriguing result, Bales et al (2006) showed that soluble Aβ binds to ChT-1, resulting in impairment of cholinergic neurotransmission in PDAPP mice. Treatment with m266 quickly restored hippocampal ACh release to wild-type levels and restored the impaired habituation response observed in PDAPP mice to wild-type levels.
Many patients with AD exhibit cerebral amyloid angiopathy (CAA), which refers to amyloid deposits in the cerebral vasculature. CAA is observed in murine AD models (Calhoun et al, 1999), and in some cases treatment with anti-Aβ antibodies increases the extent of CAA-associated microhemorrhages. DeMattos et al (2001) demonstrated that antibody binding to CAA-associated plaques correlated with CAA-associated hemorrhage. 3D6 and 10D5 that target the N terminus of Aβ, but not m266, which targets the central domain, bind CAA-associated plaques and increase the extent of microhemorrhage in the PDAPP AD mouse model (Racke et al, 2005). Separately, it was shown that treatment of Tg2576 mice with RN2286, which targets the C-terminal domain of Aβ and binds plaque (Wilcock et al, 2004a), results in CAA-associated microhemorrhage (Wilcock et al, 2004b). The results described above (Carty et al, 2006) suggest that use of deglycosylated anti-Aβ antibodies for passive immunization may be expected to reduce the incidence of CAA-associated microhemorrhage.
Both active and passive anti-Aβ immunization strategies have moved into clinical development for the treatment of AD. The first trial, involving immunization of preaggregated synthetic Aβ1−42 (AN1742; Wyeth/Elan) in combination with the adjuvant QS-21, was terminated after patients developed meningoencephalitis (Gilman et al, 2005; Orgogozo et al, 2003). Post-mortem analysis of patients revealed findings similar to those observed after immunization of PDAPP mice, including reduction in plaque load, association of collapsed plaques with microglia, and small hemorrhages (Ferrer et al, 2004; Nicoll et al, 2003). Further characterization suggested that changes in formulation between the Phase 1 and Phase 2 studies were associated in a shift in the patient T-cell response from Th2 to the proinflammatory Th1 response to the C terminus of Aβ1−42 (Pride et al, 2008). A second clinical trial using a new immunogen consisting the N-terminal domain of Aβ and a protein conjugate (ACC-001; Wyeth/Elan) is currently recruiting for Phase 2 studies (www.clinicaltrials.gov). Three of the anti-Aβ antibodies (3D6, RN1219, and m266) described above are also in clinical development. The furthest advanced is bapineuzumab (humanized 3D6; Elan/Wyeth), which is in Phase 3, whereas LY2062430 (humanized m266; Eli Lilly) is in Phase 2 and PF04360365 (humanized RN2286; Pfizer) is currently in Phase 1.
Both aptamer and siRNA approaches to Aβ neutralization/reduction have been described, although the work is at an earlier stage than the antibody-based programs. Ylera et al (2002) reported an aptamer selected against immobilized Aβ1−40 that binds to fibrils consisting Aβ1−40 with fairly high affinity (∼40 nM). As yet, no functional data regarding, for example, fibril dissociation, have been reported. Aβ peptide is produced by the sequential cleavage of the APP by β- and γ-secretases. APP transgenic mice with β-secretase (BACE1) deficiency are rescued from memory deficits (Ohno et al, 2004), suggesting that reducing Aβ load by inhibiting BACE1 may be a viable therapeutic approach. Treatment of APP mice with anti-BACE1 siRNA via a lentiviral expression system reduced the level of Aβ1−42 in the hippocampus. In addition, the siRNA-treated mice performed similarly to nontransgenic control mice in the Morris water maze (Singer et al, 2005).
Multiple Sclerosis: Anti-Interleukin-23
Multiple sclerosis is a chronic, progressive, demyelinating disease of the CNS. MS affects over 400, 000 people in the United States and may affect 2.5 million people worldwide. The pathological hallmark of the disease is white matter lesions associated with focal infiltration of mononuclear cells with variable degrees of demyelination, axonal loss, and gliosis. The susceptibility to MS is thought to be multifactorial and it is generally considered to be an autoimmune Th1-mediated disease directed against myelin antigens. Interleukin-23 (IL-23) is a member of the interleukin-12 (IL-12) family of proteins, which are predominant cytokines for triggering Th1-mediated (autoimmune) responses. IL-23 and IL-12 are heterodimeric proteins that contain an identical p40 subunit and a unique p19 (IL-23) or p35 (IL-12) subunit (Figure 7). Both IL-12 and IL-23 are produced by macrophages and dendritic cells and both cytokines affect T cells by binding to heterodimeric receptors. IL-12 acts primarily on naive T cells and promotes their differentiation into Th-1 cells that produce interferon-γ (IFN-γ). IL-23 acts primarily on Th-17 cells and is thought to stimulate their proliferation and production of IL-17 (reviewed in McKenzie et al, 2006).
Interleukin-23 is the more relevant p40-containing cytokine for experimental autoimmune encephalomyelitis (EAE) (animal model of multiple sclerosis). Clinical EAE score over time (days) for wild-type controls (filled black circles), p40−/− (filled green diamonds; IL-12/IL-23 knockout), p35−/− (closed red triangles; IL-12 knockout), and p19−/− (closed blue squares; IL-23 knockout) mice. (From: Cua et al, Nature 421:744–748, 2003).
A large amount of data have been generated linking IL-23 activity to autoimmune disease in the CNS. Mice with the p40 or the p19 subunits knocked out are highly resistant to experimental autoimmune encephalomyelitis (EAE), the mouse model for MS, whereas p35 knockout mice are, if anything, more susceptible to the disease (Figure 7; Cua et al, 2003). Moreover, intracerebral, but not intravenous, injection of IL-23 gene transfer vectors partially rescued disease susceptibility. Together, these data suggest that CNS-generated IL-23 but not IL-12 is important for EAE development. This hypothesis is supported by data showing that the IFN-γ is not required for EAE and that interleukin-17 (IL-17) knockout mice are also resistant (Ferber et al, 1996). Furthermore, Langrish et al (2005) showed that in vitro generated IL-23-dependent CD4+ T cells are highly pathogenic and essential for the establishment of CNS autoimmunity. This same pattern of resistance was seen in these knockout mice for other autoimmune disease models for rheumatoid arthritis (Murphy et al, 2003) and inflammatory bowel disease (Yen et al, 2006).
Antibody experiments validate IL-23 as a therapeutic target for MS. Neutralizing antibodies against IL-17 can ameliorate mouse EAE (Hofstetter et al, 2005) and anti-p19 (IL-23-selective) antibodies can prevent EAE induction and even reverse established disease (Chen et al, 2006). Furthermore, anti-p40 (IL-12/IL-23) antibodies have shown efficacy whether administered before or after onset of disease in a relatively new Callithrix jacchus (nonhuman primate marmoset) EAE model (‘tHart et al, 2008) and have been localized to lesions in the brain (Brok et al, 2002). In humans, overexpression of p40 mRNA (van Boxel-Dezaire et al, 1999) and p19 mRNA (Li et al, 2007) is observed in the CNS of MS patients and lower baseline levels of p40 mRNA predict clinical responsiveness to IFN-β treatment (van Boxel-Dezaire et al, 2000).
Anti-p40 antibodies, which eliminate both IL-23 and IL-12 activity, have been clinically validated for other autoimmune diseases. CNTO-1275 (ustekinumab), an anti-p40 mAb developed by Centocor Corp., has shown efficacy in both Phase 1 and Phase 2 trials for psoriasis. In the Phase 2 trial, 81% of patients receiving four weekly 90 mg doses achieved at least 75% improvement in their psoriasis, as measured by the Psoriasis Area Severity Index (PASI 75; Krueger et al, 2007). In addition, 52% achieved PASI 90, and 20% achieved PASI 100 (total skin clearance). Another anti-p40 antibody, ABT-874, showed similar results, with 90% of patients who received four weekly 200 mg subcutaneous injections achieving PASI 75 (Kimball et al, 2008). ABT-874 has also been reported to show efficacy in a Phase 2 trial for Crohn's disease (Mannon et al, 2004). Both antibodies were well tolerated in these short-term trials. It remains to be seen if these drugs will have efficacy against MS. So far one trial with CNTO-1275 in MS was conducted but the results have not been reported.
Aside from antibodies, short, stabilized anti-IL-12/23 aptamers have been reported (Burmeister et al, 2006). Several of these aptamers bind with low nanomolar affinities and inhibit IL-23- and IL-12-dependent STAT3 activation on activated human primary T cells. One of these aptamers has been truncated to 36 nucleotides and still retains activity. Thus, it shows great potential for development into a therapeutic for autoimmune diseases, including MS. However, because IL-12 is critical for immune responses, especially to intracellular pathogens (Altare et al, 1998) and not critical for the development of mouse EAE, it may be preferable to specifically target IL-23 over IL-12. To that end, aptamers selective for IL-23 have been recently developed (Pendergrast et al, 2006). One of these is able to inhibit IL-23-dependent IFN-γ release from primary human T cells over 100-fold more efficiently than IL-12-dependent IFN-γ release from the same type of cells. This same aptamer can also completely inhibit human IL-23-dependent release of IL-17 from mouse splenocyte cells (data not shown).
Huntington's Disease: Huntingtin Inhibition
Huntington's disease is an inherited neurodegenerative disorder, caused by expansion of the CAG repeat region within the huntingtin gene (The Huntington's Disease Collaborative Research Group, 1993). Clinical symptoms are characterized by involuntary movement, cognitive decline, and behavioral abnormalities; underlying these symptoms are dysfunction and degeneration of striatal and cortical neurons. Despite the identification of the mutated gene 15 years ago, there is no disease-modifying therapy available to the patient population. Functional studies on the mutated huntingtin protein and its molecular interactions with other genes and proteins have implicated huntingtin in multiple critical cellular processes including transcriptional regulation, axonal transport, apoptosis, and cell signaling. Transgenic mice expressing the mutant human huntingtin gene with expanded CAG repeat regions recapitulate molecular, cellular, and behavioral facets of Huntington's disease, providing essential systems for studying pathological mechanisms and testing therapeutic candidates (Mangiarini et al, 1996; Slow et al, 2003). However, to date, huntingtin has represented a nondrugable target and the key molecular partners in pathogenesis remain under question. Studies in mouse models with regulatable transgenes (Yamamoto et al, 2000) and virally delivered RNAi (Harper et al, 2005; Rodriguez-Lebron et al, 2005) have demonstrated that downmodulating huntingtin represents an important strategy with potential therapeutic benefit. Both unconjugated and cholesterol-conjugated siRNAs targeting huntingtin have been shown to be bioactive in vivo. Ge et al (2006; unpublished data) reported neuronal uptake of labeled siRNAs in the rodent striatum as well as huntingtin mRNA suppression after intrastriatal siRNA infusion. In a recent report, a cholesterol-conjugated siRNA was demonstrated to ameliorate neuropathology and behavioral abnormalities in a mouse model of Huntington's disease after intrastriatal administration (DiFiglia et al, 2007). Importantly, these studies have demonstrated that complete downmodulation of mutant huntingtin is not required for improvement of neuropathology or behavioral deficits; partial downmodulation of mutant huntingtin appears to be sufficient for significant benefit. In a recent report (McBride et al, 2008), shRNA expression systems that resulted in very high levels of two antisense RNAs appeared to be toxic to the mouse brain, whereas a third sequence that resulted in much lower levels of antisense RNA but similar silencing was well tolerated. Although the generalizability of these results remains to be determined, these observations illustrate the challenges of a gene therapy approach where consistency and predictability of shRNA expression levels can be problematic. In contrast, direct CNS delivery of synthetic siRNAs provides the needed degree of control over drug levels. McBride et al (2008) also report that the mouse brain can tolerate >50% reduction of huntingtin mRNA for at least 4 months, suggesting that the partial downmodulation of huntingtin that appears sufficient for significant benefit based on mouse models also is likely to be well tolerated. Both siRNA and antisense approaches are being pursued to develop drug candidates for the treatment of Huntington's disease, with direct CNS delivery. These therapeutic strategies represent novel and promising approaches that have been enabled by a unique feature of siRNA and antisense molecules—disease intervention at the mRNA level to reduce the level of a pathogenic protein, in the absence of an identified interaction to inhibit.
Parkinson's Disease: α-Synuclein Inhibition
Parkinson's disease is a neurodegenerative disorder characterized by resting tremor, rigidity, bradykinesia, and postural instability. The primary cells affected are dopaminergic nigrostriatal neurons. Current therapies primarily focus on dopamine replacement and alleviate symptoms but do not alter the underlying progression of disease. The recent clinical trial of GDNF as a disease-modifying therapeutic candidate did not provide sufficient benefit to continue clinical development. However, this trial provided an important landmark for direct intraparenchymal delivery of CNS therapeutics and for evaluation of a disease-modifying drug candidate. SNCA (α-synuclein) has been linked genetically to Parkinson's disease in familial Parkinson's disease, not only in the context of mutant SNCA (Polymeropoulos et al, 1997) but also with respect to overexpression of wild-type SNCA resulting from SNCA locus multiplication (Singleton et al, 2003; Chartier-Harlin et al, 2004; Ibáñez et al, 2004). Recently, the important discovery that SNCA is a susceptibility gene for sporadic Parkinson's disease provided further support for SNCA as a key target for therapeutic intervention (Maraganore et al, 2006). Taken together, these reports suggest that SNCA downmodulation represents a useful potential strategy for the treatment of Parkinson's disease, and that partial downmodulation of SNCA would be sufficient for therapeutic benefit. Farrer et al (2007; unpublished data) recently reported SNCA mRNA suppression after direct CNS infusion of siRNA-targeting SNCA. As with huntingtin, disease intervention at the SNCA mRNA level represents a novel and promising approach that is enabled by direct CNS delivery of RNA therapeutic modalities for reduction of a pathogenic protein.
Pain Management: Nerve Growth Factor and Artemin
Chronic pain represents a debilitating condition that severely impacts quality of life. Unlike acute pain, which has a protective role and is transient, chronic pain, such as neuropathic pain, persists and serves no useful purpose. Current therapies, predominantly consisting of small molecules targeting neurotransmitter receptors and ion channels, are inadequate in most patients and palliative rather than curative, with side effects representing significant limitations. The molecular pathways underlying chronic pain are complex and continue to be elucidated. The recent discovery that neurotrophic factors such as NGF and artemin are important in chronic pain suggests that these pathways may serve as novel intervention points for disease-modifying therapy. Although NGF was originally identified as a key neurotrophin required for the survival of pain sensory neurons, subsequent studies implicated excessive NGF in inflammatory pain as well as possibly neuropathic pain (Lewin et al, 1994; Woolf et al, 1994; McMahon et al, 1995; Koltzenburg et al, 1999). NGF levels rise dramatically in damaged or inflamed tissue, and enhanced retrograde transport of NGF occurs in sensory neurons, leading to increased levels of substance P and CGRP, which can contribute to hyperalgesia. The pivotal role of NGF in mediating inflammatory hyperalgesia was subsequently established in animal models of chronic inflammation with TrkA–IgG fusion protein and anti-NGF antiserum. On the basis of these and other data, a clinical trial was conducted with RI624, a humanized anti-NGF antibody, for the treatment of osteoarthritis pain.
Another strategy for the treatment of chronic pain is supplementation with artemin, a member of the GDNF family. Like GDNF, artemin supports the survival of cultured sensory neurons (Baloh et al, 1998a, 1998b). However, the distribution pattern of artemin's receptor, GFRα3, is relatively restricted, being expressed predominantly in adult rodent and human nociceptive sensory neurons (Orozco et al, 2001; Yang et al, 2006), suggesting that artemin would specifically modulate the pain sensory system in the adult. This was established in the rat spinal nerve ligation model where behavioral, neurochemical, and histochemical changes associated with neuropathic pain were reversed with systemic administration of artemin (Gardell et al, 2003). More recently, expression of GFRα3 immunoreactivity has also been reported in other tissues, including a subset of epithelial cells in the digestive and reproductive systems; the functional consequences of this expression remain to be determined (Yang et al, 2006). In contrast to small molecule drugs targeting individual ion channels or neurotransmitter receptors, these protein-based therapeutic strategies represent novel and promising approaches for the treatment of chronic pain that have the potential to impact multiple facets of pathology.
SUMMARY AND CONCLUSIONS
Therapeutic antibodies and oligonucleotide therapeutics such as antisense, siRNA, and aptamers represent new classes of drugs to inhibit ‘nondrugable’ targets, including the emerging class of protein–protein interaction targets. They represent excellent tools to validate targets by knocking out gene expression at the level of mRNA (antisense, RNAi, and siRNA) or protein (antibodies and aptamers). These novel therapeutic modalities (vs small molecules) have the distinct advantage of disrupting protein–protein interaction but have some limitations in their ADME properties with regard to limited oral absorption and their limited ability to cross the blood–brain barrier. Their utility for effective treatment of CNS disorders is reviewed in the context of their limited access of relevant targets outside the blood–brain barrier (eg Aβ neutralization approaches for the treatment of AD; pain targets such as NGF and artemin in the periphery), addressing diseases with a compromised blood–brain barrier such as MS (anti-IL-23 approaches) or direct delivery of the therapeutic to brain using implantable delivery devices (inhibition of huntingtin and SNCA for the treatment of HD and PD, respectively). Broader application of these novel therapeutic modalities for the treatment of neurological and psychiatric disorders is envisioned as new technologies are developed to overcome the current limitations.
References
Altare F, Durandy A, Lammas D, Emile JF, Lamhamedi S, Le Deist F et al (1998). Impairment of mycobacterial immunity in human interleukin-12 receptor deficiency. Science 280: 1432–1435.
Alter J, Lou F, Rabinowitz A, Yin H, Rosenfeld J, Wilton SD et al (2006). Systemic delivery of morpholino oligonucleotide restores dystrophin expression bodywide and improves dystrophic pathology. Nat Med 12: 175–177.
Aramburu J, Yaffe MB, Lopez-Rodriguez C, Cantley LC, Hogan PG, Rao A (1999). Affinity-driven peptide selection of an NFAT inhibitor more selective than cyclosporin A. Science 285: 2129–2133.
Auf der Maur A, Tissot K, Barberis A (2004). Antigen-independent selection of intracellular stable antibody frameworks. Methods 34: 215–224.
Bacskai BJ, Kajdasz ST, McLellan ME, Games D, Seubert P, Schenk D et al (2002). Non-Fc-mediated mechanisms are involved in clearance of amyloid-beta in vivo by immunotherapy. J Neurosci 22: 7873–7878.
Bales KR, Tzavara ET, Wu S, Wade MR, Bymaster FP, Paul SM et al (2006). Cholinergic dysfunction in a mouse model of Alzheimer disease is reversed by an anti-A beta antibody. J Clin Invest 116: 825–832.
Baloh RH, Gorodinsky A, Golden JP, Tansey MG, Keck CL, Popescu NC et al (1998a). GFRalpha3 is an orphan member of the GDNF/neurturin/persephin receptor family. Proc Natl Acad Sci USA 95: 5801–5806.
Baloh RH, Tansey MG, Lampe PA, Fahrner TJ, Enomoto H, Simburger KS et al (1998b). Artemin, a novel member of the GDNF ligand family, supports peripheral and central neurons and signals through the GFRalpha3-RET receptor complex. Neuron 21: 1291–1302.
Bard F, Cannon C, Barbour R, Burke RL, Games D, Grajeda H et al (2000). Peripherally administered antibodies against amyloid beta-peptide enter the central nervous system and reduce pathology in a mouse model of Alzheimer disease. Nat Med 6: 916–919.
Bartlett DW, Davis ME (2007). Effect of siRNA nuclease stability on the in vitro and in vivo kinetics of siRNA-mediated gene silencing. Biotechnol Bioeng 97: 909–921.
Boado RJ, Zhang Y, Zhang Y, Xia CF, Pardridge WM (2007). Fusion antibody for Alzheimer's disease with bidirectional transport across the blood–brain barrier and Aβ fibril disaggregation. Bioconjugate Chem 18: 447–455.
Bock LC, Griffin LC, Latham JA, Vermaas EH, Toole JJ (1992). Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature 355: 564–566.
Brok HP, van Meurs M, Blezer E, Schantz A, Peritt D, Treacy G et al (2002). Prevention of experimental autoimmune encephalomyelitis in common marmosets using an anti-IL-12p40 monoclonal antibody. J Immunol 169: 6554–6563.
Bumcrot D, Manoharan M, Kotelianski V, Sah DWY (2006). RNAi therapeutics: a potential new class of pharmaceutical drugs. Nat Chem Biol 2: 711–719.
Burmeister PE, Lewis SD, Simva RF, Preiss JR, Horwitz LR, Pendergrast PS et al (2005). Direct in vitro selection of a 2′-O-methyl aptamer to VEGF. Chem Biol 12: 25–33.
Burmeister PE, Wang P, Killough JR, Lewis SD, Horwitz LR, Ferguson A et al (2006). 2′-Deoxy purine, 2′-O-methyl pyrimidine (dRmY) aptamers as candidate therapeutics. Oligonucleotides 16: 337–351.
Calhoun ME, Burgermeister P, Phinney AL, Stalder M, Tolnay M, Wiederhold KH et al (1999). Neuronal overexpression of mutant amyloid precursor protein results in prominent deposition of cerebrovascular amyloid. Proc Natl Acad Sci USA 96: 14088–14093.
Carrier M, Menasche P, Levy JH, Newman MF, Taylor KM, Haverich A et al (2006). Inhibition of complement activation by pexelizumab reduces death in patients undergoing combined aortic valve replacement and coronary artery bypass surgery. J Thorac Cardiovasc Surg 131: 266–277.
Carty NC, Wilcock DM, Rosenthal A, Grimm J, Pons J, Ronan V et al (2006). Intracranial administration of deglycosylated C-terminal-specific anti-Abeta antibody efficiently clears amyloid plaques without activating microglia in amyloid-depositing transgenic mice. J Neuroinflammation 3: 11.
Chan R, Gilbert M, Marshal K, Marsh N, Epstein D (2006). Co-expression of anti-NFkappaB RNA aptamers and siRNAs leads to maximal suppression of NFkappaB activity in mammalian cells. Nucleic Acids Res 34: e36.
Chan MY, Rusconi CP, Alexander JH, Tonkens RM, Harrington RA, Becker RC (2008). A randomized, repeat dose, pharmacodynamic, and safety study of an antidote-controlled factor IXa inhibitor. J Thromb Haemost 6: 789–796.
Chapman JA, Beckey C (2006). Pegaptanib: a novel approach to ocular neovascularization. Ann Pharmacother 40: 1322–1326.
Chartier-Harlin MC, Kachergus J, Roumier C, Mouroux V, Douay X, Lincoln S et al (2004). Alpha-synuclein locus duplication as a cause of familial Parkinson's disease. Lancet 364: 1167–1169.
Chen Y, Langrish CL, McKenzie B, Joyce-Shaikh B, Stumhofer JS, McClanahan T et al (2006). Anti-IL-23 therapy inhibits multiple inflammatory pathways and ameliorates autoimmune encephalomyelitis. J Clin Invest 116: 1317–1326.
Choung S, Kim YJ, Kim S, Park HO, Choi YC (2006). Chemical modification of siRNAs to improve serum stability without loss of efficacy. Biochem Biophys Res Commun 342: 919–927.
Cload ST, McCauley TG, Keefe AD, Healy JM, Wilson C (2006). Properties of therapeutic aptamers. In: Klussmann S (ed). The Aptamer Handbook. Wiley-VCH Verlag GmbH & Co. KGaA: Weinheim. pp 363–416.
Cossum PA, Sasmor H, Dellinger D, Truong L, Cummins L, Owens SR et al (1993). Disposition of the 14C-labeled phosphorothioate oligonucleotide ISIS 2105 after intravenous administration to rats. J Pharmacol Exp Ther 267: 1181–1190.
Cossum PA, Truong L, Owens SR, Markham PM, Shea JP, Crooke ST (1994). Pharmacokinetics of a 14C-labeled phosphorothioate oligonucleotide, ISIS 2105, after intradermal administration to rats. J Pharmacol Exp Ther 269: 89–94.
Crooke ST (2004). Antisense strategies. Curr Mol Med 4: 465–487.
Cua DJ, Sherlock J, Chen Y, Murphy CA, Joyce B, Seymour B et al (2003). Interleukin-23 rather than interleukin-12 is the critical cytokine for autoimmune inflammation of the brain. Nature 421: 744–748.
Daugherty AL, Mrsny RJ (2006). Formulation and delivery issues for monoclonal antibody therapeutics. Adv Drug Deliv Rev 58: 686–706.
DeMattos RB, Bales KR, Cummings DJ, Dodart JC, Paul SM, Holtzman DM (2001). Peripheral anti-A beta antibody alters CNS and plasma A beta clearance and decreases brain A beta burden in a mouse model of Alzheimer's disease. Proc Natl Acad Sci USA 98: 8850–8855.
DiFiglia M, Sena-Esteves M, Chase K, Sapp E, Pfister E, Sass M et al (2007). Therapeutic silencing of mutant huntingtin with siRNA attenuates striatal and cortical neuropathology and behavioral deficits. Proc Natl Acad Sci USA 104: 17204–17209.
Dodart JC, Bales KR, Gannon KS, Greene SJ, DeMattos RB, Mathias C et al (2002). Immunization reverses memory deficits without reducing brain Abeta burden in Alzheimer's disease model. Nat Neurosci 5: 452–457.
Dorn G, Patel S, Wotherspoon G, Hemmings-Mieszczak M, Barclay J, Natt FJ et al (2004). siRNA relieves chronic neuropathic pain. Nuc Acids Res 32: e49.
Eaton BEL, Gold L, Hicke BJ, Janjic N, Jucker FM, Sebesta DP et al (1997). Post-SELEX combinatorial optimization of aptamers. Bioorg Med Chem 5: 1087–1096.
Evaluate Pharma (2008). http://www.evaluatepharma.com/default.htm.
Ferber IA, Brocke S, Taylor-Edwards C, Ridgway W, Dinisco C, Steinman L et al (1996). Mice with a disrupted IFN-gamma gene are susceptible to the induction of experimental autoimmune encephalomyelitis (EAE). J Immunol 156: 5–7.
Ferrer I, Boada Rovira M, Sanchez Guerra L, Rey MJ, Costa-Jussa F (2004). Neuropathology and pathogenesis of encephalitis following amyloid-beta immunization in Alzheimer's disease. Brain Pathol 14: 11–20.
Filpula D (2007). Antibody engineering and modification technologies. Biomol Eng 24: 201–215.
Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391: 806–811.
Games D, Adams D, Alessandrini R, Barbour R, Berthelette P, Blackwell C et al (1995). Alzheimer-type neuropathology in transgenic mice overexpressing V717F beta-amyloid precursor protein. Nature 373: 523–527.
Gardell LR, Wang R, Ehrenfels C, Ossipov MH, Rossomando AJ, Miller S et al (2003). Multiple actions of systemic artemin in experimental neuropathy. Nat Med 9: 1383–1389.
Gilbert JC, DeFeo-Fraulini T, Hutabarat RM, Horvath CJ, Merlino PG, Marsh NH et al (2007). First-in-human evaluation of anti von Willebrand factor therapeutic aptamer ARC1779 in healthy volunteers. Circulation 116: 2678–2686.
Gilman S, Koller M, Black RS, Jenkins L, Griffith SG, Fox NC et al (2005). Clinical effects of Abeta immunization (AN1792) in patients with AD in an interrupted trial. Neurology 64: 1553–1562.
Gold L, Polisky B, Uhlenbeck OC, Yarus M (1995). Diversity of oligonucleotide functions. Annu Rev Biochedm 64: 763–797.
Granholm AC, Backman C, Bloom F, Ebendal T, Gerhardt GA, Hoffer B et al (1994). NGF and anti-transferrin receptor antibody conjugate: short and long-term effects on survival of cholinergic neurons in intraocular septal transplants. J Pharmacol Exp Ther 268: 448–459.
Hammond SM, Boettcher S, Caudy AA, Kobayashi R, Hannon GJ (2001). Argonaute2, a link between genetic and biochemical analyses of RNAi. Science 293: 1146–1150.
Harper SQ, Staber PD, He X, Eliason SL, Martins IH, Mao Q et al (2005). RNA interference improves motor and neuropathological abnormalities in a Huntington's disease mouse model. Proc Natl Acad Sci USA 102: 5820–5825.
Hartman RE, Izumi Y, Bales KR, Paul SM, Wozniak DF, Holtzman DM (2005). Treatment with an amyloid-beta antibody ameliorates plaque load, learning deficits, and hippocampal long-term potentiation in a mouse model of Alzheimer's disease. J Neurosci 25: 6213–6220.
Hassani Z, Lemkine GF, Erbacher P, Palmier K, Alfama G, Giovannangeli C et al (2005). Lipid-mediated siRNA delivery down-regulates exogenous gene expression in the mouse brain at picomolar levels. J Gene Med 7: 198–207.
Healy JM, Lewis SD, Kurz M, Boomer RM, Thompson KM, Wilson C et al (2004). Pharmacokinetics and biodistribution of novel aptamer compositions. Pharm Res 21: 2234–2246.
Henry S, Stecker K, Brooks D, Monteith D, Conklin B, Bennett CF (2000). Chemically modified oligonucleotides exhibit decreased immune stimulation in mice. J Pharmacol Exp Ther 292: 468–479.
Hofstetter HH, Ibrahim SM, Koczan D, Kruse N, Weishaupt A, Toyka KV et al (2005). Therapeutic efficacy of IL-17 neutralization in murine experimental autoimmune encephalomyelitis. Cell Immunol 237: 123–130.
Holcomb L, Gordon MN, McGowan E, Yu X, Benkovic S, Jantzen P et al (1998). Accelerated Alzheimer-type phenotype in transgenic mice carrying both mutant amyloid precursor protein and presenilin 1 transgenes. Nat Med 4: 97–100.
Holliger P, Manzke O, Span M, Hawkins R, Fleischmann B, Qinghua B et al (1999). Carcinoembryonic antigen (CEA)-specific T-cell activation in colon carcinoma induced by anti-CD3 x anti-CEA bispecific diabodies and B7 x anti-CEA bispecific fusion proteins. Cancer Res 59: 2909–2916.
Holliger P, Hudson PJ (2005). Engineered antibody fragments and the rise of single domains. Nat Biotechnol 23: 1126–1136.
Hornung V, Guenthner-Biller M, Bourquin C, Ablasser A, Schlee M, Uematsu S et al (2005). Sequence-specific potent induction of IFN-alpha by short interfering RNA in plasmacytoid dendritic cells through TLR7. Nat Med 11: 263–270.
Huhalov A, Chester KA (2004). Engineered single chain antibody fragments for radioimmunotherapy. Q J Nucl Med Mol Imaging 48: 279–288.
Ibáñez P, Bonnet AM, Débarges B, Lohmann E, Tison F, Pollak P et al (2004). Causal relation between alpha-synuclein gene duplication and familial Parkinson's disease. Lancet 364: 1169–1171.
Jackson AL, Burchard J, Schelter J, Chau BN, Cleary M, Lim L et al (2006). Widespread siRNA ‘off-target’ transcript silencing mediated by seed region sequence complementarity. RNA 12: 1179–1187.
James W (2000). Aptamers. In: Meyers R (ed). Encyclopedia of Analytical Chemistry. Wiley: Chichester. pp 4848–4871.
Jellinek D, Green LS, Bell C, Lynott CK, Gill N, Vargeese C et al (1995). Potent 2′-amino-2′-deoxypyrimidine RNA inhibitors of basic fibroblast growth factor. Biochemistry 34: 11363–11372.
Jenison RD, Jennings SD, Walker DW, Bargatze RF, Parma D (1998). Oligonucleotide inhibitors of P-selectin-dependent neutrophil-platelet adhesion. Antisense Nucleic Acid Drug Dev 8: 265–279.
Judge AD, Sood V, Shaw JR, Fang D, McClintock K, MacLachlan I (2005). Sequence-dependent stimulation of the mammalian innate immune response by synthetic siRNA. Nat Biotechnol 23: 457–462.
Kimball AB, Gordon KB, Langley RG, Menter A, Chartiash EK, Valdes J (2008). Safety and efficacy of ABT-874, a fully human interleukin 12/23 monoclonal antibody, in the treatment of moderate to severe chronic plaque psoriasis: results of a randomized, placebo-controlled, phase 2 trial. Arch Dermatol 144: 200–207.
Koltzenburg M, Bennett DL, Shelton DL, McMahon SB (1999). Neutralization of endogenous NGF prevents the sensitization of nociceptors supplying inflamed skin. Eur J Neurosci 11: 1698–1704.
Krueger GG, Langley RG, Leonardi C, Yeilding N, Guzzo C, Wang Y et al (2007). A human interleukin-12/23 monoclonal antibody for the treatment of psoriasis. N Engl J Med 356: 580–592.
Kumar P, Lee SK, Shankar P, Manjunath N (2006). A single siRNA suppresses fatal encephalitis induced by two different flaviviruses. PLoS Med 3: e96.
Kumar P, Wu H, McBride JL, Jung KE, Kim MH, Davidson BL et al (2007). Transvascular delivery of small interfering RNA to the central nervous system. Nature 448: 39–43.
Lacerra G, Sierakowska H, Carestia C, Fucharoen S, Summerton J, Weller D et al (2000). Restoration of hemoglobin A synthesis in erythroid cells from peripheral blood of thalassemic patients. Proc Natl Acad Sci USA 97: 9591–9596.
Lackey CA, Press OW (2002). A biomimetic pH-responsive polymer directs endosomal release and intracellular delivery of an endocytosed antibody complex. Bioconjug Chem 13: 996–1001.
Lang AE, Gill S, Patel NK, Lozano A, Nutt JG, Penn R et al (2006). Randomized controlled trial of intraputamenal glial cell line-derived neurotrophic factor infusion in Parkinson disease. Ann Neurol 59: 459–466.
Langrish CL, Chen Y, Blumenschein WM, Mattson J, Basham B, Sedgwick JD et al (2005). IL-23 drives a pathogenic T cell population that induces autoimmune inflammation. J Exp Med 201: 233–240.
Lewin GR, Rueff A, Mendell LM (1994). Peripheral and central mechanisms of NGF-induced hyperalgesia. Eur J Neurosci 6: 1903–1912.
Li Y, Chu N, Hu A, Gran B, Rostami A, Zhang GX (2007). Increased IL-23p19 expression in multiple sclerosis lesions and its induction in microglia. Brain 130: 490–501.
Lichtlen P, Auf der Maur A, Barberis A (2002). Target validation through protein-domain knockout-applications of intracellularly stable single-chain antibodies. Targets 1: 37–44.
Lillo AM, Sun C, Gao C, Ditzel H, Parrish J, Gauss CM et al (2004). A human single-chain antibody specific for integrin alpha3beta1 capable of cell internalization and delivery of antitumor agents. Chem Biol 11: 897.
Lim SR, Hertel KJ (2001). Modulation of survival motor neuron pre-mRNA splicing by inhibition of alternative 3′ splice site pairing. J Biol Chem 276: 45476–45483.
Lin Y, Qiu Q, Gill SC, Jayasena SD (1994). Modified RNA sequence pools for in vitro selection. Nucleic Acids Res 22: 5229–5234.
Lo AS, Zhu Q, Marasco WA (2008). Intracellular antibodies (intrabodies) and their therapeutic potential. Handbook Exp Pharmacol 181: 343–373.
Lonberg N (2005). Human antibodies from transgenic animals. Nat Biotechnol 23: 1117–1125.
Lu QL, Rabinowitz A, Chen YC, Yokota T, Yin H, Alter J et al (2005). Systemic delivery of antisense oligoribonucleotide restores dystrophin expression in body-wide skeletal muscles. Proc Natl Acad Sci USA 102: 198–203.
Luo MC, Zhang DQ, Ma SW, Huang YY, Shuster SJ, Porreca F et al (2005). An efficient intrathecal delivery of small interfering RNA to the spinal cord and peripheral neurons. Mol Pain 1: 29.
Madocsai C, Lim SR, Geib T, Lam BJ, Hertel KJ (2005). Correction of SMN2 Pre-mRNA splicing by antisense U7 small nuclear RNAs. Mol Ther 12: 1013–1022.
Mangiarini L, Sathasivam K, Mahal A, Mott R, Seller M, Bates GP (1996). Instability of highly expanded CAG repeats in mice transgenic for the Huntington's disease mutation. Nat Genet 15: 197–200.
Mann CJ, Honeyman K, Cheng AJ, Ly T, Lloyd F, Fletcher S et al (2001). Antisense-induced exon skipping and synthesis of dystrophin in the mdx mouse. Proc Natl Acad Sci USA 98: 42–47.
Mannon PJ, Fuss IJ, Mayer L, Elson CO, Sandborn WJ, Present D et al (2004). Anti-interleukin-12 antibody for active Crohn's disease. N Engl J Med 351: 2069–2079.
Maraganore DM, de Andrade M, Elbaz A, Farrer MJ, Ioannidis JP, Krüger R, et al, Genetic Epidemiology of Parkinson's Disease (GEO-PD) Consortium (2006). Collaborative analysis of alpha-synuclein gene promoter variability and Parkinson disease. JAMA 296: 661–670.
Matranga C, Tomari Y, Shin C, Bartel DP, Zamore PD (2005). Passenger-strand cleavage facilitates assembly of siRNA into Ago2-containing RNAi enzyme complexes. Cell 123: 607–620.
McBride JL, Boudreau RL, Harper SQ, Staber PD, Monteys AM, Martins I et al (2008). Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi. Proc Natl Acad Sci USA 105: 5868–5873.
McKenzie BS, Kastelein RA, Cua DJ (2006). Understanding the IL-23-IL-17 immune pathway. Trends Immunol 27: 17–23.
Mclear JA, Lebrecht D, Messer A, Wolfgang WJ (2008). Combinational approach of intrabody with enhanced Hsp70 expression addresses multiple pathologies in a fly model of Huntington's disease. FASEB J 22: 2003–2011.
McMahon SB, Bennett DL, Priestley JV, Shelton DL (1995). The biological effects of endogenous nerve growth factor on adult sensory neurons revealed by a trkA–IgG fusion molecule. Nat Med 1: 774–780.
McNamara JO, Andrechek ER, Wang Y, Viles KD, Rempel RE, Gilboa E et al (2006). Cell type-specific delivery of siRNAs with aptamer-siRNA chimeras. Nat Biotechnol 24: 1005–1015.
McNamara JO, Kolonias D, Pastor F, Mittler RS, Chen L, Giangrande PH et al (2008). Multivalent 4-1BB binding aptamers costimulate CD8+ T cells and inhibit tumor growth in mice. J Clin Invest 118: 376–386.
Murphy CA, Langrish CL, Blumenschein W, McClanahan T, Kastelein RA, Sedgwick JD et al (2003). Divergent pro- and antiinflammatory roles for IL-23 and IL-12 in joint autoimmune inflammation. J Exp Med 198: 1951–1957.
Nicoll JA, Wilkinson D, Holmes C, Steart P, Markham H, Weller RO (2003). Neuropathology of human Alzheimer disease after immunization with amyloid-beta peptide: a case report. Nat Med 9: 448–452.
Ninichuk V, Clauss S, Kulkarni O, Schmid H, Segerer S, Radomska E et al (2008). Late onset of Ccl2 blockade with the Spiegelmer mNOX-E36-3′PEG prevents glomerulosclerosis and improves glomerular filtration rate in db/db mice. Am J Pathol 172: 628–637.
Ng EW, Shima DT, Calias P, Cunningham Jr ET, Guyer DR, Adamis AP (2006). Pegaptanib, a targeted anti-VEGF aptamer for ocular vascular disease. Nat Rev Drug Discov 5: 123–132.
Ohno M, Sametsky EA, Younkin LH, Oakley H, Younkin SG, Citron M et al (2004). BACE1 deficiency rescues memory deficits and cholinergic dysfunction in a mouse model of Alzheimer's disease. Neuron 41: 27–33.
Orgogozo JM, Gilman S, Dartigues JF, Laurent B, Puel M, Kirby LC et al (2003). Subacute meningoencephalitis in a subset of patients with AD after Abeta42 immunization. Neurology 61: 46–54.
Orozco OE, Walus L, Sah DW, Pepinsky RB, Sanicola M (2001). GFRalpha3 is expressed predominantly in nociceptive sensory neurons. Eur J Neurosci 13: 2177–2182.
Pagratis NC, Bell C, Chang YF, Jennings S, Fitzwater T, Jellinek D et al (1997). Potent 2′-amino-, and 2′-fluoro-2′-deoxyribonucleotide RNA inhibitors of keratinocyte growth factor. Nat Biotechnol 15: 68–73.
Patel NK, Bunnage M, Plaha P, Svendsen CN, Heywood P, Gill SS (2005). Intraputamenal infusion of glial cell line-derived neurotrophic factor in PD: a two-year outcome study. Arch Neurol 57: 298–302.
Pendergrast PS, Marsh JM, Grate DH, Healy JM, Stanton M (2005). Nucleic acid aptamers for target validation and therapeutic applications. J BioM Tech 16: 224–234.
Pendergrast PS, Thompson KM, Ferguson A, Killough J, Horwitz L, Lagasse HAD et al (2006). Aptamers that discriminate between IL-23 and IL-12 are specific inhibitors of IL-23 activity in vitro. GTC Bio 4th Annual Conference on Cytokines and Inflammation Jan 31 2006 San Diego California, pp 109–111.
Pich EM, Epping-Jordan M (1998). Transgenic mice in drug dependence research. Ann Med 30: 390–396.
Pietras K, Ostman A, Sjoquist M, Buch-dunger E, Reed RK, Heldin CH et al (2001). Inhibition of platelet-derived growth factor receptors reduces interstitial hypertension and increases transcapillary transport in tumors. Cancer Res 61: 2929–2934.
Pietras K, Rubin K, Sjoblom T, Buchdunger E, Sjoquist M, Heldin CH et al (2002). Inhibition of PDGF receptor signaling in tumor stroma enhances antitumor effect of chemotherapy. Cancer Res 62: 5476–5484.
Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A et al (1997). Mutation in the alpha-synuclein gene identified in families with Parkinson's disease. Science 276: 2045–2047.
Porreca F, Lai J, Bian D, Wegert S, Ossipov MH, Eglen RM et al (1999). A comparison of the potential role of the tetrodotoxin-insensitive sodium channels, PN3/SNS and NaN/SNS2, in rat models of chronic pain. Proc Natl Acad Sci USA 96: 7640–7644.
Porritt MJ, Batchelor PE, Howells DW (2005). Inhibiting BDNF expression by antisense oligonucleotide infusion causes loss of nigral dopaminergic neurons. Exp Neurol 192: 226–234.
Pride M, Seubert P, Grundman M, Hagen M, Eldridge J, Black RS (2008). Progress in the active immunotherapeutic approach to Alzheimer's disease: clinical investigations into AN1792-associated meningoencephalitis. Neurodegener Dis 5: 194–196.
Racke MM, Boone LI, Hepburn DL, Parsadainian M, Bryan MT, Ness DK et al (2005). Exacerbation of cerebral amyloid angiopathy-associated microhemorrhage in amyloid precursor protein transgenic mice by immunotherapy is dependent on antibody recognition of deposited forms of amyloid beta. J Neurosci 25: 629–636.
Rand TA, Petersen S, Du F, Wang X (2005). Argonaute2 cleaves the anti-guide strand of siRNA during RISC activation. Cell 123: 621–629.
Rhodes A, Deakin A, Spaull J, Coomber B, Aitken A, Life P et al (2000). The generation and characterization of antagonist RNA aptamers to human oncostatin M. J Biol Chem 275: 28555–28561.
Rhodes A, Smithers N, Chapman T, Parsons S, Rees S (2001). The generation and characterisation of antagonist RNA aptamers to MCP-1. FEBS Lett 506: 85–90.
Rodriguez-Lebron E, Denovan-Wright EM, Nash K, Lewin AS, Mandel RJ (2005). Intrastriatal rAAV-mediated delivery of anti-huntingtin shRNAs induces partial reversal of disease progression in R6/1 Huntington's disease transgenic mice. Mol Ther 12: 618–633.
Ruckman J, Green LS, Beeson J, Waugh S, Gillette WL, Henninger DD et al (1998). 2′-Fluoropyrimidine RNA-based aptamers to the 165-amino acid form of vascular endothelial growth factor (VEGF165). Inhibition of receptor binding and VEGF-induced vascular permeability through interactions requiring the exon 7-encoded domain. J Biol Chem 273: 20556–20567.
Rusconi CP, Scardino E, Layzer J, Pitoc GA, Ortel TL, Monroe D et al (2002). RNA aptamers as reversible antagonists of coagulation factor IXa. Nature 419: 90–94.
Sakai A, Asada M, Seno N, Suzuki H (2007). Involvement of neural cell adhesion molecule signaling in glial cell line-derived neurotrophic factor-induced analgesia in a rat model of neuropathic pain. Pain (originally published online 29 October 2007, at www.iasp-pain.org ).
Salahpour A, Medvedev IO, Beaulieu JM, Gainedinov RR, Caron MG (2007). Local knockdown of genes in the brain using small interfering RNA: a phenotypic comparison with knockout animals. Biol Psychiatry 61: 65–69.
Schenk D, Barbour R, Dunn W, Gordon G, Grajeda H, Guido T et al (1999). Immunization with amyloid-beta attenuates Alzheimer-disease-like pathology in the PDAPP mouse. Nature 400: 173–177.
Seiwert SD, Stines Nahreini T, Aigner S, Ahn NG, Uhlenbeck OC (2000). RNA aptamers as pathway-specific MAP kinase inhibitors. Chem Biol 7: 833–843.
Selkoe DJ (2001). Alzheimer's disease: genes, proteins, and therapy. Physiol Rev 81: 741–766.
Selkoe DJ, Schenk D (2003). Alzheimer's disease: molecular understanding predicts amyloid-based therapeutics. Annu Rev Pharmacol Toxicol 43: 545–584.
Senn JJ, Burel S, Henry SP (2005). Non-CpG-containing antisense 2′-methoxyethyl oligonucleotides activate a proinflammatory response independent of Toll-like receptor 9 or myeloid differentiation factor 88. J Pharmacol Exp Ther 314: 972–979.
Seubert P, Vigo-Pelfrey C, Esch F, Lee M, Dovey H, Davis D et al (1992). Isolation and quantification of soluble Alzheimer's beta-peptide from biological fluids. Nature 359: 325–327.
Shamah SM, Healy JM, Cload ST (2008). Complex target SELEX. Acc Chem Res 41: 130–138.
Singer O, Marr RA, Rockenstein E, Crews L, Coufal NG, Gage FH et al (2005). Targeting BACE1 with siRNAs ameliorates Alzheimer disease neuropathology in a transgenic model. Nat Neurosci 8: 1343–1349.
Singleton AB, Farrer M, Johnson J, Singleton A, Hague S, Kachergus J et al (2003). alpha-Synuclein locus triplication causes Parkinson's disease. Science 302: 841.
Slevin JT, Gash DM, Smith CD, Gerhardt GA, Kryscio R, Chebrolu H et al (2006). Unilateral intraputaminal glial cell line-derived neurotrophic factor in patients with Parkinson disease: response to 1 year each of treatment and withdrawal. Neurosurg Focus 20: E1.
Slow EJ, van Raamsdonk J, Rogers D, Coleman SH, Graham RK, Deng Y et al (2003). Selective striatal neuronal loss in a YAC128 mouse model of Huntington disease. Hum Mol Genet 12: 1555–1567.
Smith RA, Miller TM, Yamanaka K, Monia BP, Condon TP, Hung G et al (2006). Antisense oligonucleotide therapy for neurodegenerative disease. J Clin Invest 116: 2290–2296.
Spencer BJ, Verma IM (2007). Targeted delivery of proteins across the blood–brain barrier. Proc Natl Acad Sci USA 104: 7594–7599.
Tan PH, Yang LC, Shih HC, Lan KC, Cheng JT (2005). Gene knockdown with intrathecal siRNA of NMDA receptor NR2B subunit reduces formalin-induced nociception in the rat. Gene Therapy 12: 59–66.
Thakker DR, Natt F, Husken D, Maier R, Muller M, van der Putten H et al (2004). Neurochemical and behavioral consequences of widespread gene knockdown in the adult mouse brain by using nonviral RNA interference. Proc Natl Acad Sci USA 101: 17270–17275.
Thakker DR, Natt F, Husken D, van der Putten H, Maier R, Hoyer D et al (2005). siRNA-mediated knockdown of the serotonin transporter in the adult mouse brain. Mol Psychiatry 10: 782–789.
‘tHart BA, Hintzen RQ, Laman JD (2008). Preclinical assessment of therapeutic antibodies against human CD40 and human interleukin-12/23p40 in a nonhuman primate model of multiple sclerosis. Neurodegener Dis 5: 38–52.
Tuerk C, Gold L (1990). Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science 249: 505–510.
Van Boxel-Dezaire AH, Hoff SC, van Oosten BW, Verweij CL, Drager AM, Ader HJ et al (1999). Decreased interleukin-10 and increased interleukin-12p40 mRNA are associated with disease activity and characterize different disease stages in multiple sclerosis. Ann Neurol 45: 695–703.
Van Boxel-Dezaire AH, van Trigt-Hoff SC, Killestein J, Schrijver HM, van Houwelingen JC, Polman CH et al (2000). Contrasting responses to interferon beta-1b treatment in relapsing-remitting multiple sclerosis: does baseline interleukin-12p35 messenger RNA predict the efficacy of treatment? Ann Neurol 48: 313–322.
Wahlestedt C, Pich EM, Koob GF, Yee F, Heilig M (1993). Modulation of anxiety and neuropeptide Y-Y1 receptors by antisense oligodeoxynucleotides. Science 259: 528–531.
Wang HM, He M (2007). The rapid discovery of engineered antibodies. IDrugs 10: 562–565.
Wang YL, Liu W, Wada E, Murata M, Wada K, Kanazawa I (2005). Clinico-pathological rescue of a model mouse of Huntington's disease by siRNA. Neuroscience Res 53: 241–249.
Wark KL, Hudson PJ (2006). Latest technologies for the enhancement of antibody affinity. Adv Drug Deliv Rev 58: 657–670.
Watson SR, Chang YF, O'Connell D, Wiegand L, Ringquist S, Parma DH (2000). Anti-L-selectin aptamers: binding characteristics, pharmacokinetic parameters, and activity against an intravascular target in vivo. Antisense Nucleic Acid Drug Dev 10: 63–75.
Wiegand TW, Williams PB, Dreskin SC, Jouvin MH, Kinet JP, Tasset D (1996). High-affinity oligonucleotide ligands to human IgE inhibit binding to Fc epsilon receptor I. J Immunol 157: 221–230.
Wilcock DM, Rojiani A, Rosenthal A, Levkowitz G, Subbarao S, Alamed J et al (2004a). Passive amyloid immunotherapy clears amyloid and transiently activates microglia in a transgenic mouse model of amyloid deposition. J Neurosci 24: 6144–6151.
Wilcock DM, Rojiani A, Rosenthal A, Subbarao S, Freeman MJ, Gordon MN et al (2004b). Passive immunotherapy against Abeta in aged APP-transgenic mice reverses cognitive deficits and depletes parenchymal amyloid deposits in spite of increased vascular amyloid and microhemorrhage. J Neuroinflammation 1: 24–34.
Wilson C, Keefe AD (2006). Building oligonucleotide therapeutics using non-natural chemistries. Curr Opin Chem Biol 10: 607–614.
Woolf CJ, Safieh-Garabedian B, Ma QP, Crilly P, Winter J (1994). Nerve growth factor contributes to the generation of inflammatory sensory hypersensitivity. Neurosci 62: 327–331.
Xia CF, Zhang Y, Zhang Y, Boado RJ, Pardridge WM (2007). Intravenous siRNA of brain cancer with receptor targeting and avidin-biotin technology. Pharma Res 24: 2309–2316.
Yamamoto A, Lucas JJ, Hen R (2000). Reversal of neuropathology and motor dysfunction in a conditional model of Huntington's disease. Cell 101: 57–66.
Yang X, Gorenstein DG (2004). Progress in thioaptamer development. Curr Drug Targets 5: 705–715.
Yang C, Hutto D, Sah DWY (2006). Distribution of GDNF family receptor α3 and RET in rat and human non-neural tissues. J Mol Histol 37: 69–77.
Yen D, Cheung J, Scheerens H, Poulet F, McClanahan T, Mckenzie B et al (2006). IL-23 is essential for T cell-mediated colitis and promotes inflammation via IL-17 and IL-6. J Clin Invest 116: 1310–1316.
Ylera F, Lurz R, Erdmann VA, Furste JP (2002). Selection of RNA aptamers to the Alzheimer's disease amyloid peptide. Biochem Biophys Res Commun 290: 1583–1588.
Acknowledgements
We thank Eric Gornstein for helpful research. The authors are full-time employees of Archemix Corporation, a company focused on therapeutic aptamers (Errol B De Souza, Sharon T Cload, and P Shannon Pendergrast) and Alnylam Pharmaceuticals, a company focused on siRNA development (Dinah WY Sah). In addition, Errol B De Souza is on the following boards of directors or scientific advisory boards (Bionomics Ltd, IDEXX Laboratories, National Institute of Mental Health Board of Scientific Counselors, Palatin Technologies, and Targacept Inc.).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
De Souza, E., Cload, S., Pendergrast, P. et al. Novel Therapeutic Modalities to Address Nondrugable Protein Interaction Targets. Neuropsychopharmacol 34, 142–158 (2009). https://doi.org/10.1038/npp.2008.115
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/npp.2008.115
Keywords
This article is cited by
-
Intravenously Administered Novel Liposomes, DCL64, Deliver Oligonucleotides to Cerebellar Purkinje Cells
The Cerebellum (2019)
-
Oncogenic protein interfaces: small molecules, big challenges
Nature Reviews Cancer (2014)
-
Deciphering the Roles of Trehalose and Hsp104 in the Inhibition of Aggregation of Mutant Huntingtin in a Yeast Model of Huntington’s Disease
NeuroMolecular Medicine (2014)
-
Principles and application of antibody libraries for infectious diseases
Biotechnology Letters (2014)
-
Tricyclic pyrone compounds prevent aggregation and reverse cellular phenotypes caused by expression of mutant huntingtin protein in striatal neurons
BMC Neuroscience (2009)