Abstract
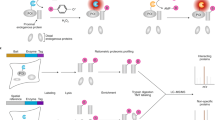
Here we describe the protein interaction platform assay, a method for identifying interacting proteins in Saccharomyces cerevisiae. This assay relies on the reovirus scaffolding protein μNS, which forms large focal inclusions in living cells. When a query protein is fused to μNS and potential interaction partners are fused to a fluorescent reporter, interactors can be identified by screening for yeast that display fluorescent foci.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Fields, S. & Song, O. Nature 340, 245–246 (1989).
Remy, I. & Michnick, S.W. Proc. Natl. Acad. Sci. USA 96, 5394–5399 (1999).
Miller, C.L. et al. Mol. Cell. Proteomics 6, 1027–1038 (2007).
Nguyen, C.L., Eichwald, C., Nibert, M.L. & Munger, K. J. Virol. 81, 13533–13543 (2007).
Broering, T.J. et al. J. Virol. 79, 6194–6206 (2005).
Ménard, R., Sansonetti, P., Parsot, C. & Vasselon, T. Cell 79, 515–525 (1994).
Ogawa, M., Suzuki, T., Tatsuno, I., Abe, H. & Sasakawa, C. Mol. Microbiol. 48, 913–931 (2003).
Niebuhr, K. et al. Mol. Microbiol. 38, 8–19 (2000).
Hachani, A. et al. Microbes Infect. 10, 260–268 (2008).
Page, A.L., Sansonetti, P. & Parsot, C. Mol. Microbiol. 43, 1533–1542 (2002).
Parsot, C. et al. Mol. Microbiol. 56, 1627–1635 (2005).
Page, A.L., Fromont-Racine, M., Sansonetti, P., Legrain, P. & Parsot, C. Mol. Microbiol. 42, 1133–1145 (2001).
Slagowski, N.L., Kramer, R.W., Morrison, M.F., LaBaer, J. & Lesser, C.F. PLoS Pathog. 4, e9 (2008).
Alto, N.M. et al. Cell 124, 133–145 (2006).
Darwin, K.H., Robinson, L.S. & Miller, V.L. J. Bacteriol. 183, 1452–1454 (2001).
Ehrbar, K., Friebel, A., Miller, S.I. & Hardt, W.D. J. Bacteriol. 185, 6950–6967 (2003).
Huh, W.K. et al. Nature 425, 686–691 (2003).
Marsischky, G. & LaBaer, J. Genome Res. 14, 2020–2028 (2004).
Alberti, S., Gitler, A.D. & Lindquist, S. Yeast 24, 913–919 (2007).
Walhout, A.J. & Vidal, M. Methods 24, 297–306 (2001).
Weiss, D.S. et al. J. Bacteriol. 181, 508–520 (1999).
Goehring, N.W., Gonzalez, M.D. & Beckwith, J. Mol. Microbiol. 61, 33–45 (2006).
Uetz, P. et al. Nature 403, 623–627 (2000).
Datsenko, K.A. & Wanner, B.L. Proc. Natl. Acad. Sci. USA 97, 6640–6645 (2000).
Acknowledgements
We thank S. Alberti and S. Lindquist (Whitehead Institute, Massachusetts Institute of Technology) for providing the pAG destination clones; A. Gray, K. Fixen and M. Goldberg (Massachusetts General Hospital, Harvard Medical School) for providing plasmids pNG162 and pDSW206 and antibodies to IcsA and isocitrate dehydrogenase; J. Heindl (Massachusetts General Hospital, Harvard Medical School) for providing the IpgB2 W62A construct; T. Hao, D. Hill and M. Vidal (Dana Farber Cancer Institute, Harvard Medical School) for assistance in designing primers to create the S. flexneri Gateway entry clones and for providing the Gateway-compatible Y2H vectors; and R. Levy and C. Koser (Massachusetts General Hospital, Harvard Medical School) for cloning and expressing S. typhimurium effectors in yeast. We also thank the US National Institute of Allergy and Infectious Diseases and the J. Craig Venter Institute for supplying the S. typhimurium Gateway entry clones. Partial support for this work was provided by US National Institutes of Health grants R56 AI067445 to M.L.N. and R01 AI064285 to C.F.L., and by a Charles E. Culpeper Medical Scholarship from the Rockefeller Brothers Fund and Goldman Philanthropic Partnerships to C.F.L.
Author information
Authors and Affiliations
Contributions
A.M.S., M.F.M. and A.O.A. performed and analyzed the experiments. A.M.S., M.L.N. and C.F.L. designed the experiments. M.L.N. and C.F.L. wrote the manuscript.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–7 and Supplementary Tables 1 and 2 (PDF 7447 kb)
Rights and permissions
About this article
Cite this article
Schmitz, A., Morrison, M., Agunwamba, A. et al. Protein interaction platforms: visualization of interacting proteins in yeast. Nat Methods 6, 500–502 (2009). https://doi.org/10.1038/nmeth.1337
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth.1337
This article is cited by
-
Structures of autoinhibited and polymerized forms of CARD9 reveal mechanisms of CARD9 and CARD11 activation
Nature Communications (2019)
-
Intermediate filaments enable pathogen docking to trigger type 3 effector translocation
Nature Microbiology (2016)