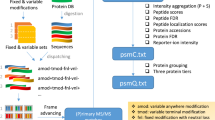
Abstract
Shotgun proteomics uses liquid chromatography–tandem mass spectrometry to identify proteins in complex biological samples. We describe an algorithm, called Percolator, for improving the rate of confident peptide identifications from a collection of tandem mass spectra. Percolator uses semi-supervised machine learning to discriminate between correct and decoy spectrum identifications, correctly assigning peptides to 17% more spectra from a tryptic Saccharomyces cerevisiae dataset, and up to 77% more spectra from non-tryptic digests, relative to a fully supervised approach.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Eng, J.K., McCormack, A.L. & Yates, J.R. III. J. Am. Soc. Mass Spectrom. 5, 976–989 (1994).
Perkins, D.N., Pappin, D.J.C., Creasy, D.M. & Cottrell, J.S. Electrophoresis 20, 3551–3567 (1999).
MacCoss, M.J., Wu, C.C. & Yates, J.R. III. Anal. Chem. 74, 5593–5599 (2002).
Keller, A., Nezvizhskii, A.I., Kolker, E. & Aebersold, R. Anal. Chem. 74, 5383–5392 (2002).
Moore, R.E., Young, M.K. & Lee, T.D. J. Am. Soc. Mass Spectrom. 13, 378–386 (2002).
Peng, J., Elias, J.E., Thoreen, C.C., Licklider, L.J. & Gygi, S.P. J. Proteome Res. 2, 43–50 (2003).
Anderson, D.C., Li, W., Payan, D.G. & Noble, W.S. J. Proteome Res. 2, 137–146 (2003).
Boser, B.E., Guyon, I.M. & Vapnik, V.N. A training algorithm for optimal margin classifiers. in 5th Annual ACM Workshop on COLT (ed. Haussler, D.) 144–152 (ACM Press, Pittsburgh, Pennsylvania, USA, 1992).
Storey, J.D. & Tibshirani, R. Proc. Natl. Acad. Sci. USA 100, 9440–9445 (2003).
Tabb, D.L., McDonald, W.H. & Yates, J.R. III. J. Proteome Res. 1, 21–26 (2002).
Washburn, M.P., Wolters, D. & Yates, J.R. III. Nat. Biotechnol. 19, 242–247 (2001).
Acknowledgements
This work was funded by US National Institutes of Health grants P41 RR011823 and R01 EB007057.
Author information
Authors and Affiliations
Contributions
M.J.M. came up with the initial idea to use decoy PSMs as negative examples. L.K. and W.S.N. came up with the idea to use a support vector machine using semi-supervised learning. L.K. implemented Percolator and performed computational experiments. J.W. provided machine learning expertise. J.D.C. performed initial proof-of-concept experiment and provided mass spectrometry expertise. W.S.N., L.K. and M.J.M. wrote the article.
Corresponding author
Supplementary information
Supplementary Text and Figures
Supplementary Figures 1–4, Supplementary Tables 1 and 2, Supplementary Methods, Supplementary Data (PDF 1393 kb)
Rights and permissions
About this article
Cite this article
Käll, L., Canterbury, J., Weston, J. et al. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat Methods 4, 923–925 (2007). https://doi.org/10.1038/nmeth1113
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nmeth1113
This article is cited by
-
Characterization of a nuclear transport factor 2-like domain-containing protein in Plasmodium berghei
Malaria Journal (2024)
-
Kinase inhibitor pulldown assay (KiP) for clinical proteomics
Clinical Proteomics (2024)
-
Chemoproteomic development of SLC15A4 inhibitors with anti-inflammatory activity
Nature Chemical Biology (2024)
-
Magnetic transferrin nanoparticles (MTNs) assay as a novel isolation approach for exosomal biomarkers in neurological diseases
Biomaterials Research (2023)
-
Combination of automated sample preparation and micro-flow LC–MS for high-throughput plasma proteomics
Clinical Proteomics (2023)