Announcements
-
-
-

A call for ecology papers
Open for submissions -
Advertisement
-
-

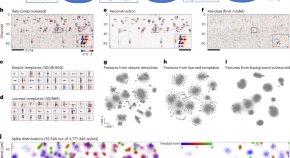
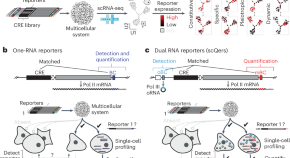
Quantitative profiling of regulatory DNA activity at single-cell resolution
We developed a two-pronged strategy to functionally probe the enormous repertoire of noncoding DNA within genomes. Our approach markedly improved signal-to-noise ratio and successfully intersected single-cell genomics with reporter assays. The result delivers a multiplex and highly quantitative readout of regulatory sequences’ activity in dynamic and multicellular systems.
-
-
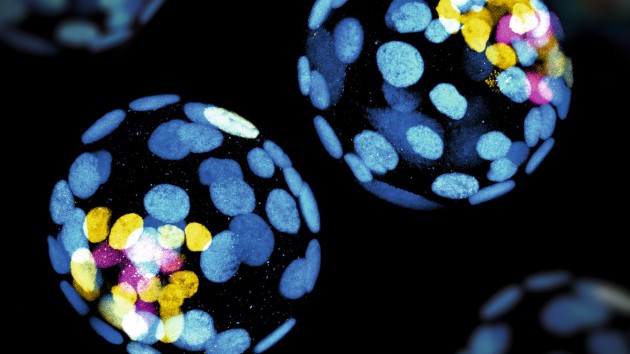
Method of the Year 2023: Methods for modeling development
Methods for modeling development is our Method of the Year 2023, for the remarkable insights that recent methodological advances have enabled in our understanding of the molecular mechanisms of human embryogenesis.
Trending - Altmetric
-
Molecular pixelation: spatial proteomics of single cells by sequencing
-
Protein–ligand structure prediction
-
Multiplex profiling of developmental cis-regulatory elements with quantitative single-cell expression reporters
-
Scalable telomere-to-telomere assembly for diploid and polyploid genomes with double graph