Abstract
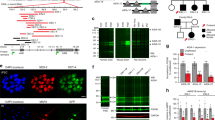
Cayman ataxia is a recessive congenital ataxia restricted to one area of Grand Cayman Island1,2. Comparative mapping suggested that the locus on 19p13.3 associated with Cayman ataxia might be homologous to the locus on mouse chromosome 10 associated with the recessive ataxic mouse mutant jittery. Screening genes in the region of overlap identified mutations in a novel predicted gene in three mouse jittery alleles, including the first mouse mutation caused by an Alu-related (B1 element) insertion. We found two mutations exclusively in all individuals with Cayman ataxia. The gene ATCAY or Atcay encodes a neuron-restricted protein called caytaxin. Caytaxin contains a CRAL-TRIO motif common to proteins that bind small lipophilic molecules. Mutations in another protein containing a CRAL-TRIO domain, alpha-tocopherol transfer protein (TTPA), cause a vitamin E–responsive ataxia. Three-dimensional protein structural modeling predicts that the caytaxin ligand is more polar than vitamin E. Identification of the caytaxin ligand may help develop a therapy for Cayman ataxia.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others

References
Johnson, W.G., Murphy, M., Murphy, W.I. & Bloom, A.D. Recessive congenital cerebellar disorder in a genetic isolate: CPD type VII? Neurology 28, 352–353 (1978).
Brown, L., Mueller, M. & Benke, P.J. A non-progressive cerebellar ataxia on Grand Cayman Island. Neurology 34, 273 (1984).
Nystuen, A., Benke, P.J., Merren, J., Stone, E.M. & Sheffield, V.C. A cerebellar ataxia locus identified by DNA pooling to search for linkage disequilibrium in an isolated population from the Cayman Islands. Hum. Mol. Genet. 5, 525–531 (1996).
Kapfhamer, D. et al. The neurological mouse mutations jittery and hesitant are allelic and map to the region of mouse chromosome 10 homologous to 19p13.3. Genomics 35, 533–538 (1996).
Kazazian, H.H. Jr. & Moran, J.V. The impact of L1 retrotransposons on the human genome. Nat. Genet. 19, 19–24 (1998).
Liu, H.X., Cartegni, L., Zhang, M.Q. & Krainer, A.R. A mechanism for exon skipping caused by nonsense or missense mutations in BRCA1 and other genes. Nat. Genet. 27, 55–58 (2001).
Boyd, J.M. et al. Adenovirus E1B 19 kDa and Bcl-2 proteins interact with a common set of cellular proteins. Cell 79, 341–351 (1994).
Zhang, H.M. et al. Nip21 gene expression reduces coxsackievirus B3 replication by promoting apoptotic cell death via a mitochondria-dependent pathway. Circ. Res. 90, 1251–1258 (2002).
Stocker, A., Tomizaki, T., Schulze-Briese, C. & Baumann, U. Crystal structure of the human supernatant protein factor. Structure (Camb) 10, 1533–1540 (2002).
Sha, B., Phillips, S.E., Bankaitis, V.A. & Luo, M. Crystal structure of the Saccharomyces cerevisiae phosphatidylinositol-transfer protein. Nature 391, 506–510 (1998).
Ouahchi, K. et al. Ataxia with isolated vitamin E deficiency is caused by mutations in the alpha-tocopherol transfer protein. Nat. Genet. 9, 141–145 (1995).
Drewett, P.L., Scudder, S.J. & Quitmyer, I.R. Unoccupied Islands? The Cayman Islands. in Prehistoric Settlements in the Caribbean 5–16 (Archetype Publications, London, UK, 2000).
Dietrich, W. et al. A genetic map of the mouse suitable for typing intraspecific crosses. Genetics 131, 423–447 (1992).
Puttagunta, R. et al. Comparative maps of human 19p13.3 and mouse chromosome 10 allow identification of sequences at evolutionary breakpoints. Genome Res. 10, 1369–1380 (2000).
Reese, M.G., Eeckman, F.H., Kulp, D. & Haussler, D. Improved splice site detection in Genie. J. Comput. Biol. 4, 311–323 (1997).
Burge, C. & Karlin, S. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 268, 78–94 (1997).
Herman, J.P., Wiegand, S.J. & Watson, S.J. Regulation of basal corticotropin-releasing hormone and arginine vasopressin messenger ribonucleic acid expression in the paraventricular nucleus: effects of selective hypothalamic deafferentations. Endocrinology 127, 2408–2417 (1990).
Lin, C.H. et al. Neurological abnormalities in a knock-in mouse model of Huntington's disease. Hum. Mol. Genet. 10, 137–144 (2001).
Dehal, P. et al. Human chromosome 19 and related regions in mouse: conservative and lineage-specific evolution. Science 293, 104–111 (2001).
Kelley, L.A., MacCallum, R.M. & Sternberg, M.J. Enhanced genome annotation using structural profiles in the program 3D- PSSM. J. Mol. Biol. 299, 499–520 (2000).
Acknowledgements
We thank the subjects and their family members on the Cayman Islands for their participation; K. Portman, D. Kapfhamer, J. Lee, C. Pontrello, C. Searby and D. Sufalko for technical assistance; M. Owen for the British control samples; P. Nishina and C. Avery for the sidewinder mutant mice; M. Reese for help interpreting splice site predicting program results; D. Jenne, T. Wilkie and A. Olsen for communication of unpublished results; N. Gilbert and J. Moran for discussion of B1-element insertions; D. Speert, C. Neal and A. Seasholtz for help with in situ hybridization; and the Morphology Core of the Center for Organogenesis of the University of Michigan for providing slides of mouse embryos. We thank the National Institute of Neurological Disorders and Stroke (M.B.), the March of Dimes Birth Defect Foundation (M.B.) and the government of the Cayman Islands (P.J.B.) for funding. V.C.S. is an investigator of the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The regents of the University of Michigan have filed two provisional patent applications titled 'Ataxia-associated gene and protein'.
Supplementary information
Rights and permissions
About this article
Cite this article
Bomar, J., Benke, P., Slattery, E. et al. Mutations in a novel gene encoding a CRAL-TRIO domain cause human Cayman ataxia and ataxia/dystonia in the jittery mouse. Nat Genet 35, 264–269 (2003). https://doi.org/10.1038/ng1255
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ng1255
This article is cited by
-
Novel homozygous frameshift variant in the ATCAY gene in an Iranian patient with Cayman cerebellar ataxia; expanding the neuroimaging and clinical features: a case report
BMC Medical Genomics (2023)
-
A combination of multiple autoantibodies is associated with the risk of Alzheimer’s disease and cognitive impairment
Scientific Reports (2022)
-
Degradation of Caytaxin Causes Learning and Memory Deficits via Activation of DAPK1 in Aging
Molecular Neurobiology (2019)
-
The Classification of Autosomal Recessive Cerebellar Ataxias: a Consensus Statement from the Society for Research on the Cerebellum and Ataxias Task Force
The Cerebellum (2019)
-
Systematic review of autosomal recessive ataxias and proposal for a classification
Cerebellum & Ataxias (2017)