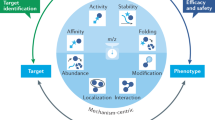
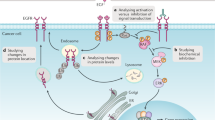
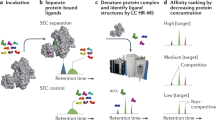
Abstract
The medical and pharmaceutical communities are facing a dire need for new druggable targets, while, paradoxically, the targets of some drugs that are in clinical use or development remain elusive. Many compounds have been found to be more promiscuous than originally anticipated, which can potentially lead to side effects, but which may also open up additional medical uses. As we move toward systems biology and personalized medicine, comprehensively determining small molecule–target interaction profiles and mapping these on signaling and metabolic pathways will become increasingly necessary. Chemical proteomics is a powerful mass spectrometry–based affinity chromatography approach for identifying proteome-wide small molecule–protein interactions. Here we will provide a critical overview of the basic concepts and recent advances in chemical proteomics and review recent applications, with a particular emphasis on kinase inhibitors and natural products.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Booth, B. & Zemmel, R. Prospects for productivity. Nat. Rev. Drug Discov. 3, 451–456 (2004).
Brown, D. Unfinished business: target-based drug discovery. Drug Discov. Today 12, 1007–1012 (2007).
Stockwell, B.R. Exploring biology with small organic molecules. Nature 432, 846–854 (2004).
Bain, J. et al. The selectivity of protein kinase inhibitors: a further update. Biochem. J. 408, 297–315 (2007).
Buchdunger, E. et al. Inhibition of the Abl protein-tyrosine kinase in vitro and in vivo by a 2-phenylaminopyrimidine derivative. Cancer Res. 56, 100–104 (1996).
Quintás-Cardama, A., Kantarjian, H. & Cortes, J. Flying under the radar: the new wave of BCR-ABL inhibitors. Nat. Rev. Drug Discov. 6, 834–848 (2007).
Zhang, J., Yang, P.L. & Gray, N.S. Targeting cancer with small molecule kinase inhibitors. Nat. Rev. Cancer 9, 28–39 (2009).
Borisy, A.A. et al. Systematic discovery of multicomponent therapeutics. Proc. Natl. Acad. Sci. USA 100, 7977–7982 (2003).
Meisner, N.C. et al. The chemical hunt for the identification of drugable targets. Curr. Opin. Chem. Biol. 8, 424–431 (2004).
Overington, J.P., Al-Lazikani, B. & Hopkins, A.L. How many drug targets are there? Nat. Rev. Drug Discov. 5, 993–996 (2006).
Lamb, J. et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science 313, 1929–1935 (2006).
Cohen, A.A. et al. Dynamic proteomics of individual cancer cells in response to a drug. Science 322, 1511–1516 (2008).
Watkins, S.M. & German, J.B. Metabolomics and biochemical profiling in drug discovery and development. Curr. Opin. Mol. Ther. 4, 224–228 (2002).
Hillenmeyer, M.E. et al. The chemical genomic portrait of yeast: uncovering a phenotype for all genes. Science 320, 362–365 (2008).
Zon, L.I. & Peterson, R.T. In vivo drug discovery in the zebrafish. Nat. Rev. Drug Discov. 4, 35–44 (2005).
Becker, F. et al. A three-hybrid approach to scanning the proteome for targets of small molecule kinase inhibitors. Chem. Biol. 11, 211–223 (2004).
Caligiuri, M. et al. MASPIT: three-hybrid trap for quantitative proteome fingerprinting of small molecule-protein interactions in mammalian cells. Chem. Biol. 13, 711–722 (2006).
Jaeger, S., Eriani, G. & Martin, F. Results and prospects of the yeast three-hybrid system. FEBS Lett. 556, 7–12 (2004).
Sadaghiani, A.M., Verhelst, S.H. & Bogyo, M. Tagging and detection strategies for activity-based proteomics. Curr. Opin. Chem. Biol. 11, 20–28 (2007).
Barglow, K.T. & Cravatt, B.F. Activity-based protein profiling for the functional annotation of enzymes. Nat. Methods 4, 822–827 (2007).
Hagenstein, M.C. & Sewald, N. Chemical tools for activity-based proteomics. J. Biotechnol. 124, 56–73 (2006).
Doucet, A., Butler, G.S., Rodriguez, D., Prudova, A. & Overall, C.M. Metadegradomics: toward in vivo quantitative degradomics of proteolytic post-translational modifications of the cancer proteome. Mol. Cell. Proteomics 7, 1925–1951 (2008).
Cuatrecasas, P., Wilchek, M. & Anfinsen, C.B. Selective enzyme purification by affinity chromatography. Proc. Natl. Acad. Sci. USA 61, 636–643 (1968).
Harding, M.W., Galat, A., Uehling, D.E. & Schreiber, S.L. A receptor for the immunosuppressant FK506 is a cis-trans peptidyl-prolyl isomerase. Nature 341, 758–760 (1989).
Crews, C.M., Collins, J.L., Lane, W.S., Snapper, M.L. & Schreiber, S.L. GTP-dependent binding of the antiproliferative agent didemnin to elongation factor 1 alpha. J. Biol. Chem. 269, 15411–15414 (1994).
Knockaert, M. et al. Intracellular targets of cyclin-dependent kinase inhibitors: identification by affinity chromatography using immobilised inhibitors. Chem. Biol. 7, 411–422 (2000).
Wilm, M. & Mann, M. Analytical properties of the nanoelectrospray ion source. Anal. Chem. 68, 1–8 (1996).
Ong, S.E. & Mann, M. Mass spectrometry-based proteomics turns quantitative. Nat. Chem. Biol. 1, 252–262 (2005).
Oda, Y., Huang, K., Cross, F.R., Cowburn, D. & Chait, B.T. Accurate quantitation of protein expression and site-specific phosphorylation. Proc. Natl. Acad. Sci. USA 96, 6591–6596 (1999).
Gygi, S.P. et al. Quantitative analysis of complex protein mixtures using isotope-coded affinity tags. Nat. Biotechnol. 17, 994–999 (1999).
Veenstra, T.D., Martinovic, S., Anderson, G.A., Pasa-Tolic, L. & Smith, R.D. Proteome analysis using selective incorporation of isotopically labeled amino acids. J. Am. Soc. Mass Spectrom. 11, 78–82 (2000).
Wilm, M., Neubauer, G. & Mann, M. Parent ion scans of unseparated peptide mixtures. Anal. Chem. 68, 527–533 (1996).
Mann, M. & Jensen, O.N. Proteomic analysis of post-translational modifications. Nat. Biotechnol. 21, 255–261 (2003).
Aebersold, R. & Mann, M. Mass spectrometry-based proteomics. Nature 422, 198–207 (2003).
Shoemaker, B.A. & Panchenko, A.R. Deciphering protein-protein interactions. Part I. Experimental techniques and databases. PLoS Comput. Biol. 3, e42 (2007).
Brehmer, D., Godl, K., Zech, B., Wissing, J. & Daub, H. Proteome-wide identification of cellular targets affected by bisindolylmaleimide-type protein kinase C inhibitors. Mol. Cell. Proteomics 3, 490–500 (2004).
Bach, S. et al. Roscovitine targets, protein kinases and pyridoxal kinase. J. Biol. Chem. 280, 31208–31219 (2005).
Rix, U. et al. Chemical proteomic profiles of the BCR-ABL inhibitors imatinib, nilotinib, and dasatinib reveal novel kinase and nonkinase targets. Blood 110, 4055–4063 (2007).
Aye, T.T. et al. Selectivity in enrichment of cAMP-dependent protein kinase regulatory subunits type I and type II and their interactors using modified cAMP affinity resins. Mol. Cell Proteomics 8, 1016–1028 (2009).
Ong, S.E. et al. Identifying the proteins to which small-molecule probes and drugs bind in cells. Proc. Natl. Acad. Sci. USA 106, 4617–4622 (2009).
Brehmer, D. et al. Cellular targets of gefitinib. Cancer Res. 65, 379–382 (2005).
Bantscheff, M. et al. Quantitative chemical proteomics reveals mechanisms of action of clinical ABL kinase inhibitors. Nat. Biotechnol. 25, 1035–1044 (2007).
Daub, H. et al. Kinase-selective enrichment enables quantitative phosphoproteomics of the kinome across the cell cycle. Mol. Cell 31, 438–448 (2008).
Zhang, Y.X. et al. AXL is a potential target for therapeutic intervention in breast cancer progression. Cancer Res. 68, 1905–1915 (2008).
Zhou, H.X., Rivas, G. & Minton, A.P. Macromolecular crowding and confinement: biochemical, biophysical, and potential physiological consequences. Annu. Rev. Biophys. 37, 375–397 (2008).
Oda, Y. et al. Quantitative chemical proteomics for identifying candidate drug targets. Anal. Chem. 75, 2159–2165 (2003).
Kaida, D. et al. Spliceostatin A targets SF3b and inhibits both splicing and nuclear retention of pre-mRNA. Nat. Chem. Biol. 3, 576–583 (2007).
Wang, G., Shang, L., Burgett, A.W., Harran, P.G. & Wang, X. Diazonamide toxins reveal an unexpected function for ornithine delta-amino transferase in mitotic cell division. Proc. Natl. Acad. Sci. USA 104, 2068–2073 (2007).
Schirle, M., Heurtier, M.A. & Kuster, B. Profiling core proteomes of human cell lines by one-dimensional PAGE and liquid chromatography-tandem mass spectrometry. Mol. Cell. Proteomics 2, 1297–1305 (2003).
Shiyama, T., Furuya, M., Yamazaki, A., Terada, T. & Tanaka, A. Design and synthesis of novel hydrophilic spacers for the reduction of nonspecific binding proteins on affinity resins. Bioorg. Med. Chem. 12, 2831–2841 (2004).
Knockaert, M. et al. p42/p44 MAPKs are intracellular targets of the CDK inhibitor purvalanol. Oncogene 21, 6413–6424 (2002).
Taunton, J., Hassig, C.A. & Schreiber, S.L. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science 272, 408–411 (1996).
Shimizu, N. et al. High-performance affinity beads for identifying drug receptors. Nat. Biotechnol. 18, 877–881 (2000).
Speers, A.E. & Cravatt, B.F. A tandem orthogonal proteolysis strategy for high-content chemical proteomics. J. Am. Chem. Soc. 127, 10018–10019 (2005).
van der Veken, P. et al. Development of a novel chemical probe for the selective enrichment of phosphorylated serine- and threonine-containing peptides. ChemBioChem 6, 2271–2280 (2005).
Fauq, A.H., Kache, R., Khan, M.A. & Vega, I.E. Synthesis of acid-cleavable light isotope-coded affinity tags (ICAT-L) for potential use in proteomic expression profiling analysis. Bioconjug. Chem. 17, 248–254 (2006).
Shimkus, M., Levy, J. & Herman, T. A chemically cleavable biotinylated nucleotide: usefulness in the recovery of protein-DNA complexes from avidin affinity columns. Proc. Natl. Acad. Sci. USA 82, 2593–2597 (1985).
Verhelst, S.H., Fonovic, M. & Bogyo, M. A mild chemically cleavable linker system for functional proteomic applications. Angew. Chem. Int. Edn Engl. 46, 1284–1286 (2007).
Fonović, M., Verhelst, S.H., Sorum, M.T. & Bogyo, M. Proteomics evaluation of chemically cleavable activity-based probes. Mol. Cell. Proteomics 6, 1761–1770 (2007).
Griffith, E.C. et al. Methionine aminopeptidase (type 2) is the common target for angiogenesis inhibitors AGM-1470 and ovalicin. Chem. Biol. 4, 461–471 (1997).
Sin, N., Meng, L., Auth, H. & Crews, C.M. Eponemycin analogues: syntheses and use as probes of angiogenesis. Bioorg. Med. Chem. 6, 1209–1217 (1998).
Bantscheff, M., Schirle, M., Sweetman, G., Rick, J. & Kuster, B. Quantitative mass spectrometry in proteomics: a critical review. Anal. Bioanal. Chem. 389, 1017–1031 (2007).
Katayama, H. & Oda, Y. Chemical proteomics for drug discovery based on compound-immobilized affinity chromatography. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 855, 21–27 (2007).
Winger, J.A., Hantschel, O., Superti-Furga, G. & Kuriyan, J. The structure of the leukemia drug imatinib bound to human quinone reductase 2 (NQO2). BMC Struct. Biol. 9, 7 (2009).
Förster, T. Intermolecular energy migration and fluorescence. Ann. Phys. 2, 55–75 (1948).
Duncan, J.S. et al. An unbiased evaluation of CK2 inhibitors by chemoproteomics: characterization of inhibitor effects on CK2 and identification of novel inhibitor targets. Mol. Cell. Proteomics 7, 1077–1088 (2008).
Ghoreschi, K., Laurence, A. & O'Shea, J.J. Selectivity and therapeutic inhibition of kinases: to be or not to be? Nat. Immunol. 10, 356–360 (2009).
Fabian, M.A. et al. A small molecule-kinase interaction map for clinical kinase inhibitors. Nat. Biotechnol. 23, 329–336 (2005).
Karaman, M.W. et al. A quantitative analysis of kinase inhibitor selectivity. Nat. Biotechnol. 26, 127–132 (2008).
Godl, K. et al. An efficient proteomics method to identify the cellular targets of protein kinase inhibitors. Proc. Natl. Acad. Sci. USA 100, 15434–15439 (2003).
Wissing, J. et al. Chemical proteomic analysis reveals alternative modes of action for pyrido[2,3-d]pyrimidine kinase inhibitors. Mol. Cell. Proteomics 3, 1181–1193 (2004).
Godl, K. et al. Proteomic characterization of the angiogenesis inhibitor SU6668 reveals multiple impacts on cellular kinase signaling. Cancer Res. 65, 6919–6926 (2005).
Wan, Y. et al. Synthesis and target identification of hymenialdisine analogs. Chem. Biol. 11, 247–259 (2004).
Kim, E. & Park, J.M. Identification of novel target proteins of cyclic GMP signaling pathways using chemical proteomics. J. Biochem. Mol. Biol. 36, 299–304 (2003).
Scholten, A. et al. Analysis of the cGMP/cAMP interactome using a chemical proteomics approach in mammalian heart tissue validates sphingosine kinase type 1-interacting protein as a genuine and highly abundant AKAP. J. Proteome Res. 5, 1435–1447 (2006).
Scholten, A., van Veen, T.A., Vos, M.A. & Heck, A.J. Diversity of cAMP-dependent protein kinase isoforms and their anchoring proteins in mouse ventricular tissue. J. Proteome Res. 6, 1705–1717 (2007).
Remsing Rix, L.L. et al. Global target profile of the kinase inhibitor bosutinib in primary chronic myeloid leukemia cells. Leukemia 23, 477–485 (2009).
Hantschel, O. et al. The Btk tyrosine kinase is a major target of the Bcr-Abl inhibitor dasatinib. Proc. Natl. Acad. Sci. USA 104, 13283–13288 (2007).
Feher, M. & Schmidt, J.M. Property distributions: differences between drugs, natural products, and molecules from combinatorial chemistry. J. Chem. Inf. Comput. Sci. 43, 218–227 (2003).
Piggott, A.M. & Karuso, P. Quality, not quantity: the role of natural products and chemical proteomics in modern drug discovery. Comb. Chem. High Throughput Screen. 7, 607–630 (2004).
Chen, J.K., Lane, W.S. & Schreiber, S.L. The identification of myriocin-binding proteins. Chem. Biol. 6, 221–235 (1999).
Adam, G.C., Vanderwal, C.D., Sorensen, E.J. & Cravatt, B.F. (−)-FR182877 is a potent and selective inhibitor of carboxylesterase-1. Angew. Chem. Int. Edn. Engl. 42, 5480–5484 (2003).
Kotake, Y. et al. Splicing factor SF3b as a target of the antitumor natural product pladienolide. Nat. Chem. Biol. 3, 570–575 (2007).
Low, W.K. et al. Inhibition of eukaryotic translation initiation by the marine natural product pateamine A. Mol. Cell 20, 709–722 (2005).
Bordeleau, M.E. et al. Stimulation of mammalian translation initiation factor eIF4A activity by a small molecule inhibitor of eukaryotic translation. Proc. Natl. Acad. Sci. USA 102, 10460–10465 (2005).
Bordeleau, M.E. et al. RNA-mediated sequestration of the RNA helicase eIF4A by Pateamine A inhibits translation initiation. Chem. Biol. 13, 1287–1295 (2006).
Low, W.K., Dang, Y., Bhat, S., Romo, D. & Liu, J.O. Substrate-dependent targeting of eukaryotic translation initiation factor 4A by pateamine A: negation of domain-linker regulation of activity. Chem. Biol. 14, 715–727 (2007).
van der Greef, J. et al. The art and practice of systems biology in medicine: mapping patterns of relationships. J. Proteome Res. 6, 1540–1559 (2007).
Schena, M., Shalon, D., Davis, R.W. & Brown, P.O. Quantitative monitoring of gene expression patterns with a complementary DNA microarray. Science 270, 467–470 (1995).
Raamsdonk, L.M. et al. A functional genomics strategy that uses metabolome data to reveal the phenotype of silent mutations. Nat. Biotechnol. 19, 45–50 (2001).
Mizuarai, S., Irie, H., Schmatz, D.M. & Kotani, H. Integrated genomic and pharmacological approaches to identify synthetic lethal genes as cancer therapeutic targets. Curr. Mol. Med. 8, 774–783 (2008).
Brehme, M. et al. Charting the molecular network of the drug target Bcr-Abl. Proc. Natl. Acad. Sci. USA 106, 7414–7419 (2009).
Rigaut, G. et al. A generic protein purification method for protein complex characterization and proteome exploration. Nat. Biotechnol. 17, 1030–1032 (1999).
Bürckstümmer, T. et al. An efficient tandem affinity purification procedure for interaction proteomics in mammalian cells. Nat. Methods 3, 1013–1019 (2006).
Glatter, T., Wepf, A., Aebersold, R. & Gstaiger, M. An integrated workflow for charting the human interaction proteome: insights into the PP2A system. Mol. Syst. Biol. 5, 237 (2009).
Kruse, U., Bantscheff, M., Drewes, G. & Hopf, C. Chemical and pathway proteomics: powerful tools for oncology drug discovery and personalized health care. Mol. Cell. Proteomics 7, 1887–1901 (2008).
Snyder, J.R. et al. Dissection of melanogenesis with small molecules identifies prohibitin as a regulator. Chem. Biol. 12, 477–484 (2005).
Zhang, Q. et al. Small-molecule synergist of the Wnt/beta-catenin signaling pathway. Proc. Natl. Acad. Sci. USA 104, 7444–7448 (2007).
Bachovchin, D.A., Brown, S.J., Rosen, H. & Cravatt, B.F. Identification of selective inhibitors of uncharacterized enzymes by high-throughput screening with fluorescent activity-based probes. Nat. Biotechnol. 27, 387–394 (2009).
Hagenstein, M.C. et al. Affinity-based tagging of protein families with reversible inhibitors: a concept for functional proteomics. Angew. Chem. Int. Edn. Engl. 42, 5635–5638 (2003).
Ge, X. & Sem, D.S. Affinity-based chemical proteomic probe for dehydrogenases: fluorescence and visible binding assays in gels. Anal. Biochem. 370, 171–179 (2007).
Acknowledgements
We thank M. Brehme for graphical assistance and our colleagues for providing information about the most current publications in press. We acknowledge financial support from the Austrian Federal Ministry for Science and Research under the GEN-AU program (GZ 200.142/1-VI/1/2006).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
G.S.-F. holds stocks of Cellzome AG.
Rights and permissions
About this article
Cite this article
Rix, U., Superti-Furga, G. Target profiling of small molecules by chemical proteomics. Nat Chem Biol 5, 616–624 (2009). https://doi.org/10.1038/nchembio.216
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.216
This article is cited by
-
In-silico target prediction by ensemble chemogenomic model based on multi-scale information of chemical structures and protein sequences
Journal of Cheminformatics (2023)
-
Oncoproteomics: insight into current proteomic technologies in cancer biomarker discovery and treatment
Journal of Proteins and Proteomics (2022)
-
Target identification of natural medicine with chemical proteomics approach: probe synthesis, target fishing and protein identification
Signal Transduction and Targeted Therapy (2020)
-
N,N-Dimethylaminopyrene as a fluorescent affinity mass tag for ligand-binding mode analysis
Scientific Reports (2020)
-
Chemical proteomics reveals target selectivity of clinical Jak inhibitors in human primary cells
Scientific Reports (2019)