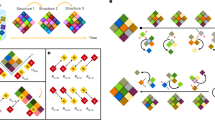
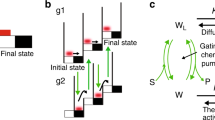
Abstract
The thermodynamic principle of cooperativity is used to drive the formation of specific macromolecular complexes during the assembly of a macromolecular machine. Understanding cooperativity provides insight into the mechanisms that govern assembly and disassembly of multicomponent complexes. Our understanding of assembly mechanisms is lagging considerably behind our understanding of the structure and function of these complexes. A significant challenge remains in tackling the thermodynamics and kinetics of the intermolecular interactions required for all cellular functions.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
References
Pawson, T. & Nash, P. Assembly of cell regulatory systems through protein interaction domains. Science 300, 445–452 (2003).
Perutz, M.F., Wilkinson, A.J., Paoli, M. & Dodson, G.G. The stereochemical mechanism of the cooperative effects in hemoglobin revisited. Annu. Rev. Biophys. Biomol. Struct. 27, 1–34 (1998).
Dill, K.A. et al. Principles of protein-folding—a perspective from simple exact models. Protein Sci. 4, 561–602 (1995).
Horovitz, A. & Fersht, A.R. Strategy for analyzing the cooperativity of intramolecular interactions in peptides and proteins. J. Mol. Biol. 214, 613–617 (1990).
Kranz, J.K. & Hall, K.B. RNA recognition by the human U1A protein is mediated by a network of local cooperative interactions that create the optimal binding surface. J. Mol. Biol. 285, 215–231 (1999).
Olson, A.J., Hu, Y.H.E. & Keinan, E. Chemical mimicry of viral capsid self-assembly. Proc. Natl. Acad. Sci. USA 104, 20731–20736 (2007).
Ackers, G.K., Johnson, A.D. & Shea, M.A. Quantitative model for gene-regulation by lambda-phage repressor. Proc. Natl. Acad. Sci. USA 79, 1129–1133 (1982).
Johnson, A.D. et al. Lambda-repressor and cro—components of an efficient molecular switch. Nature 294, 217–223 (1981).
Artavanis-Tsakonas, S., Rand, M.D. & Lake, R.J. Notch signaling: cell fate control and signal integration in development. Science 284, 770–776 (1999).
Schroeter, E.H., Kisslinger, J.A. & Kopan, R. Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature 393, 382–386 (1998).
De Strooper, B. et al. A presenilin-1-dependent gamma-secretase-like protease mediates release of Notch intracellular domain. Nature 398, 518–522 (1999).
Petcherski, A.G. & Kimble, J. LAG-3 is a putative transcriptional activator in the C-elegans Notch pathway. Nature 405, 364–368 (2000).
Nam, Y., Sliz, P., Song, L.Y., Aster, J.C. & Blacklow, S.C. Structural basis for cooperativity in recruitment of MAML coactivators to Notch transcription complexes. Cell 124, 973–983 (2006).
Del Bianco, C., Aster, J.C. & Blacklow, S.C. Mutational and energetic studies of notch1 transcription complexes. J. Mol. Biol. 376, 131–140 (2008).
Held, W.A., Ballou, B., Mizushima, S. & Nomura, M. Assembly mapping of 30 S ribosomal proteins from Escherichia coli. Further studies. J. Biol. Chem. 249, 3103–3111 (1974).
Schluenzen, F. et al. Structure of functionally activated small ribosomal subunit at 3.3 Å resolution. Cell 102, 615–623 (2000).
Wimberly, B.T. et al. The structure of the 30S ribosomal subunit. Nature 407, 327–339 (2000).
Recht, M.I. & Williamson, J.R. Central domain assembly: thermodynamics and kinetics of S6 and S18 binding to and S15-RNA complex. J. Mol. Biol. 313, 35–48 (2001).
Recht, M.I. & Williamson, J.R. RNA tertiary structure and cooperative assembly of a large ribonucleoprotein complex. J. Mol. Biol. 344, 395–407 (2004).
Dill, K.A., Fiebig, K.M. & Chan, H.S. Cooperativity in protein-folding kinetics. Proc. Natl. Acad. Sci. USA 90, 1942–1946 (1993).
Hochstrasser, M. Ubiquitin-dependent protein degradation. Annu. Rev. Genet. 30, 405–439 (1996).
Voges, D., Zwickl, P. & Baumeister, W. The 26S proteasome: a molecular machine designed for controlled proteolysis. Annu. Rev. Biochem. 68, 1015–1068 (1999).
Hirano, Y. et al. Cooperation of multiple chaperones required for the assembly of mammalian 20S proteasomes. Mol. Cell 24, 977–984 (2006).
Hirano, Y. et al. A heterodimeric complex that promotes the assembly of mammalian 20S proteasomes. Nature 437, 1381–1385 (2005).
Kusmierczyk, A.R., Kunjappu, M.J., Funakoshi, M. & Hochstrasser, M. A multimeric assembly factor controls the formation of alternative 20S proteasomes. Nat. Struct. Mol. Biol. 15, 237–244 (2008).
Le Tallec, B. et al. 20S proteasome assembly is orchestrated by two of chaperones in yeast distinct pairs and in mammals. Mol. Cell 27, 660–674 (2007).
Li, X., Kusmierczyk, A.R., Wong, P., Emili, A. & Hochstrasser, M. β-subunit appendages promote 20S proteasome assembly by overcoming an Ump1-dependent checkpoint. EMBO J. 26, 2339–2349 (2007).
Yashiroda, H. et al. Crystal structure of a chaperone complex that contributes to the assembly of yeast 20S proteasomes. Nat. Struct. Mol. Biol. 15, 228–236 (2008).
Rosenzweig, R. & Glickman, M.H. Forging a proteasome alpha-ring with dedicated proteasome chaperones. Nat. Struct. Mol. Biol. 15, 218–220 (2008).
Pollard, T.D. Regulation of actin filament assembly by Arp2/3 complex and formins. Annu. Rev. Biophys. Biomol. Struct. 36, 451–477 (2007).
Torres, F.E. et al. Enthalpy arrays. Proc. Natl. Acad. Sci. USA 101, 9517–9522 (2004).
Velazquez-Campoy, A. & Freire, E. ITC in the post-genomic era...? Priceless. Biophys. Chem. 115, 115–124 (2005).
Gracias, D.H., Tien, J., Breen, T.L., Hsu, C. & Whitesides, G.M. Forming electrical networks in three dimensions by self-assembly. Science 289, 1170–1172 (2000).
Acknowledgements
The author thanks S. Edgcomb for helpful discussions and critical reading of the manuscript, S. Blacklow for critical comments and E. Vogel of Leonardo's Basement for the photo of the large Soma cube. This work was supported by a grant from the US National Institutes of Health (GM-53757 to J.R.W.).
Author information
Authors and Affiliations
Rights and permissions
About this article
Cite this article
Williamson, J. Cooperativity in macromolecular assembly. Nat Chem Biol 4, 458–465 (2008). https://doi.org/10.1038/nchembio.102
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio.102
This article is cited by
-
Repurposing host-guest chemistry to sequester virulence and eradicate biofilms in multidrug resistant Pseudomonas aeruginosa and Acinetobacter baumannii
Nature Communications (2023)
-
Functional advantages of building nanosystems using multiple molecular components
Nature Chemistry (2023)
-
Programming conformational cooperativity to regulate allosteric protein-oligonucleotide signal transduction
Nature Communications (2023)
-
Smart polymer chemosensors: signal-amplification systems with allosterism
Polymer Journal (2021)
-
Defective internal allosteric network imparts dysfunctional ATP/substrate-binding cooperativity in oncogenic chimera of protein kinase A
Communications Biology (2021)