Abstract
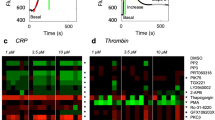
Short synthetic oligopeptides based on regions of human proteins that encompass functional motifs are versatile reagents for understanding protein signaling and interactions. They can either mimic or inhibit the parent protein's activity1,2,3,4 and have been used in drug development5. Peptide studies typically either derive peptides from a single identified protein or (at the other extreme) screen random combinatorial peptides4,6, often without knowledge of the signaling pathways targeted. Our objective was to determine whether rational bioinformatic design of oligopeptides specifically targeted to potentially signaling-rich juxtamembrane regions could identify modulators of human platelet function. High-throughput in vitro platelet function assays of palmitylated cell-permeable oligopeptides corresponding to these regions identified many agonists and antagonists of platelet function. Many bioactive peptides were from adhesion molecules, including a specific CD226-derived inhibitor of inside-out platelet signaling. Systematic screens of this nature are highly efficient tools for discovering short signaling motifs in molecular signaling pathways.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Covic, L., Misra, M., Badar, J., Singh, C. & Kuliopulos, A. Pepducin-based intervention of thrombin-receptor signaling and systemic platelet activation. Nat. Med. 8, 1161–1165 (2002).
Larkin, D. et al. ICln, a novel integrin αIIbβ3-associated protein, functionally regulates platelet activation. J. Biol. Chem. 279, 27286–27293 (2004).
Stephens, G. et al. A sequence within the cytoplasmic tail of GpIIb independently activates platelet aggregation and thromboxane synthesis. J. Biol. Chem. 273, 20317–20322 (1998).
Kamb, A. & Teng, D.H. Transdominant genetics, peptide inhibitors and drug targets. Curr. Opin. Mol. Ther. 2, 662–669 (2000).
Scarborough, R.M. et al. Barbourin. A GPIIb-IIIa-specific integrin antagonist from the venom of Sistrurus m. barbouri. J. Biol. Chem. 266, 9359–9362 (1991).
Shih, Y.P. et al. High-throughput screening of soluble recombinant proteins. Protein Sci. 11, 1714–1719 (2002).
McRedmond, J.P. et al. Integration of proteomics and genomics in platelets: a profile of platelet proteins and platelet-specific genes. Mol. Cell. Proteomics 3, 133–144 (2004).
Sowa, M.E. et al. Prediction and confirmation of a site critical for effector regulation of RGS domain activity. Nat. Struct. Biol. 8, 234–237 (2001).
Liu, J., Jackson, C.W., Gruppo, R.A., Jennings, L.K. & Gartner, T.K. The β3 subunit of the integrin αIIbβ3 regulates αIIb-mediated outside-in signaling. Blood 105, 4345–4352 (2005).
Godehardt, A.W., Hammerschmidt, S., Frank, R. & Chhatwal, G.S. Binding of α2-macroglobulin to GRAB (Protein G-related α2-macroglobulin-binding protein), an important virulence factor of group A streptococci, is mediated by two charged motifs in the DeltaA region. Biochem. J. 381, 877–885 (2004).
Kumagai, A. & Dunphy, W.G. Repeated phosphopeptide motifs in Claspin mediate the regulated binding of Chk1. Nat. Cell Biol. 5, 161–165 (2003).
Moran, N. et al. Monitoring modulators of platelet aggregation in a microtiter plate assay. Anal. Biochem. 357, 77–84 (2006).
Sun, B., Tandon, N.N., Yamamoto, N., Yoshitake, M. & Kambayashi, J. Luminometric assay of platelet activation in 96-well microplate. Biotechniques 31, 1174–1178 (2001).
Aylward, K., Meade, G., Ahrens, I., Devocelle, M. & Moran, N. A novel functional role for the highly conserved α-subunit KVGFFKR motif distinct from integrin αIIbβ3 activation processes. J. Thromb. Haemost. 4, 1804–1812 (2006).
Yamanouchi, J., Hato, T., Tamura, T. & Fujita, S. Suppression of integrin activation by the membrane-distal sequence of the integrin αIIb cytoplasmic tail. Biochem. J. 379, 317–323 (2004).
Searls, D.B. Pharmacophylogenomics: genes, evolution and drug targets. Nat. Rev. Drug Discov. 2, 613–623 (2003).
Kojima, H. et al. CD226 mediates platelet and megakaryocytic cell adhesion to vascular endothelial cells. J. Biol. Chem. 278, 36748–36753 (2003).
Chen, L. et al. The expression, regulation and adhesion function of a novel CD molecule, CD226, on human endothelial cells. Life Sci. 73, 2373–2382 (2003).
Slupsky, J.R., Cawley, J.C., Kaplan, C. & Zuzel, M. Analysis of CD9, CD32 and p67 signalling: use of degranulated platelets indicates direct involvement of CD9 and p67 in integrin activation. Br. J. Haematol. 96, 275–286 (1997).
Sun, C., Jin, B. & Liu, X. PTA1 monoclonal antibody induces human platelet aggregation and intra-cytoplasmic Ca2+ elevation. Zhonghua Xue Ye Xue Za Zhi 19, 133–137 (1998).
Shibuya, K. et al. CD226 (DNAM-1) is involved in lymphocyte function-associated antigen 1 costimulatory signal for naive T cell differentiation and proliferation. J. Exp. Med. 198, 1829–1839 (2003).
Hillmann, A.G. et al. Comparative RNA expression analyses from small-scale, single-donor platelet samples. J. Thromb. Haemost. 4, 349–356 (2006).
Puntervoll, P. et al. ELM server: a new resource for investigating short functional sites in modular eukaryotic proteins. Nucleic Acids Res. 31, 3625–3630 (2003).
Stephens, G. et al. A sequence within the cytoplasmic tail of GpIIb independently activates platelet aggregation and thromboxane synthesis. J. Biol. Chem. 273, 20317–20322 (1998).
Martin, K., Meade, G., Moran, N., Shields, D.C. & Kenny, D. A palmitylated peptide derived from the glycoprotein Ib beta cytoplasmic tail inhibits platelet activation. J. Thromb. Haemost. 1, 2643–2652 (2003).
Davey, N.E., Shields, D.C. & Edwards, R.J. SLiMDisc: short, linear motif discovery, correcting for common evolutionary descent. Nucleic Acids Res. 34, 3546–3554 (2006).
Nitabach, M.N., Llamas, D.A., Thompson, I.J., Collins, K.A. & Holmes, T.C. Phosphorylation-dependent and phosphorylation-independent modes of modulation of shaker family voltage-gated potassium channels by SRC family protein tyrosine kinases. J. Neurosci. 22, 7913–7922 (2002).
Neduva, V. et al. Systematic discovery of new recognition peptides mediating protein interaction networks. PLoS Biol. 3, e405 (2005).
Edwards, R.J. & Shields, D.C. BADASP: predicting functional specificity in protein families using ancestral sequences. Bioinformatics 21, 4190–4191 (2005).
Acknowledgements
The authors thank V. Buckley, Y. Po-Ba, E. Bernard, S. Desgranges and C. Petit for the synthesis of the peptides. This work was funded by Health Research Board, Science Foundation Ireland, and the Programme for Research in Third Level Institutions.
Author information
Authors and Affiliations
Contributions
D.C.S., D.K. and N.M. devised the experiment; R.J.E. and D.C.S. formulated the bioinformatics peptide design algorithm and performed the statistical analysis; R.J.E., N.M., D.K. and D.C.S. cowrote the paper; R.J.E. developed the bioinformatics tools and performed the peptide design and the bioinformatics searches for shared or known motifs in bioactive peptides; M.D. designed the selection rules for the synthesis of the peptides to ensure maximum integrity, stability and solubility of the corresponding palmitylated sequences and supervised the synthesis of these peptides; N.M., A.K. and E.D. developed and performed the high-throughput platelet activation assays; G.M. performed the colocalization microscopy; W.S., D.K. and N.M. designed and interpreted the platelet spreading assays; S.D.E.P. performed platelet expression analysis; M.F. provided proteomics data.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Schema of method with hypothesized mechanisms of action. (PDF 496 kb)
Supplementary Fig. 2
Example identification of peptide of interest. (PDF 607 kb)
Supplementary Fig. 3
Dose response of CD226_Q palmitylated peptide. (PDF 109 kb)
Supplementary Table 1
Statistical analysis and peptide details for high-throughput assays at two doses. (PDF 48 kb)
Supplementary Table 2
Top 20 motifs shared among peptides identified by SLiMDisc analysis. (PDF 17 kb)
Supplementary Table 3
Potential protein-protein interaction motifs present in active peptides. (PDF 21 kb)
Rights and permissions
About this article
Cite this article
Edwards, R., Moran, N., Devocelle, M. et al. Bioinformatic discovery of novel bioactive peptides. Nat Chem Biol 3, 108–112 (2007). https://doi.org/10.1038/nchembio854
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio854
This article is cited by
-
GIP_HUMAN[22–51] is a new proatherogenic peptide identified by native plasma peptidomics
Scientific Reports (2021)
-
Optimization of a MT1-MMP-targeting Peptide and Its Application in Near-infrared Fluorescence Tumor Imaging
Scientific Reports (2018)
-
Computational survey of peptides derived from disulphide-bonded protein loops that may serve as mediators of protein-protein interactions
BMC Bioinformatics (2014)
-
Shotgun Proteomic Analysis of Emiliania huxleyi, a Marine Phytoplankton Species of Major Biogeochemical Importance
Marine Biotechnology (2011)