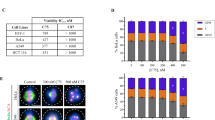
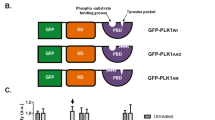
Abstract
Cell-permeable small molecules that inhibit their targets on fast timescales are powerful probes of cell-division mechanisms. Such inhibitors have been identified using phenotype-based screens with chemical libraries. However, the characteristics of compound libraries needed to effectively span cell-division phenotype space, to find probes that target different mechanisms, are not known. Here we show that a small collection of 100 diaminopyrimidines (DAPs) yields a range of cell-division phenotypes, including changes in spindle geometry, chromosome positioning and mitotic index. Monopolar mitotic spindles are induced by four inhibitors, including one that targets Polo-like kinases (Plks), evolutionarily conserved serine/threonine kinases. Using chemical inhibitors and high-resolution live-cell microscopy, we found that Plk activity is needed for the assembly and maintenance of bipolar mitotic spindles. Plk inhibition destabilizes kinetochore microtubules while stabilizing other spindle microtubules, leading to monopolar spindles. Further testing of compounds based on 'privileged scaffolds', such as the DAP scaffold, could lead to new cell-division probes and antimitotic agents.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$259.00 per year
only $21.58 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
References
Rajagopalan, H. & Lengauer, C. Aneuploidy and cancer. Nature 432, 338–341 (2004).
Mitchison, T.J. & Salmon, E.D. Mitosis: a history of division. Nat. Cell Biol. 3, E17–E21 (2001).
Inoue, S. & Salmon, E.D. Force generation by microtubule assembly/disassembly in mitosis and related movements. Mol. Biol. Cell 6, 1619–1640 (1995).
Lampson, M.A. & Kapoor, T.M. Unraveling cell division mechanisms with small-molecule inhibitors. Nat. Chem. Biol. 2, 19–27 (2006).
Jordan, M.A. & Wilson, L. Microtubules as a target for anticancer drugs. Nat. Rev. Cancer 4, 253–265 (2004).
Bettencourt-Dias, M. et al. Genome-wide survey of protein kinases required for cell cycle progression. Nature 432, 980–987 (2004).
Kittler, R. & Buchholz, F. Functional genomic analysis of cell division by endoribonuclease-prepared siRNAs. Cell Cycle 4, 564–567 (2005).
Fitzgerald, K. RNAi versus small molecules: different mechanisms and specificities can lead to different outcomes. Curr. Opin. Drug Discov. Devel. 8, 557–566 (2005).
Specht, K.M. & Shokat, K.M. The emerging power of chemical genetics. Curr. Opin. Cell Biol. 14, 155–159 (2002).
Tan, D.S. Diversity-oriented synthesis: exploring the intersections between chemistry and biology. Nat. Chem. Biol. 1, 74–84 (2005).
Lipinski, C. & Hopkins, A. Navigating chemical space for biology and medicine. Nature 432, 855–861 (2004).
Evans, B.E. et al. Methods for drug discovery: development of potent, selective, orally effective cholecystokinin antagonists. J. Med. Chem. 31, 2235–2246 (1988).
De Corte, B.L. From 4,5,6,7-tetrahydro-5-methylimidazo[4,5,1-jk](1,4)benzodiazepin-2(1H)-one (TIBO) to etravirine (TMC125): fifteen years of research on non-nucleoside Inhibitors of HIV-1 reverse transcriptase. J. Med. Chem. 48, 1689–1696 (2005).
Sorbera, L.A., Castaner, J. & Martin, L. Revaprazan hydrochloride: treatment of GERD, H+/K+-ATPase inhibitor, antiulcer drug. Drugs Future 29, 455–459 (2004).
Breault, G.A. et al. Cyclin-dependent kinase 4 inhibitors as a treatment for cancer. Part 2: identification and optimisation of substituted 2,4-bis anilino pyrimidines. Bioorg. Med. Chem. Lett. 13, 2961–2966 (2003).
Legendre, P. & Legendre, L. Numerical ecology (Elsevier, New York, 1998).
Hotha, S. et al. HR22C16: a potent small-molecule probe for the dynamics of cell division. Angew. Chem. Int. Edn. Engl. 42, 2379–2382 (2003).
Aronov, A.M. & Murcko, M.A. Toward a pharmacophore for kinase frequent hitters. J. Med. Chem. 47, 5616–5619 (2004).
Feng, B.Y., Shelat, A., Doman, T.N., Guy, R.K. & Shoichet, B.K. High-throughput assays for promiscuous inhibitors. Nat. Chem. Biol. 1, 146–148 (2005).
Burdine, L. & Kodadek, T. Target identification in chemical genetics: the (often) missing link. Chem. Biol. 11, 593–597 (2004).
Davis-Ward, R., Mook, R.A., Jr., Neeb, M.J. & Salovich, J.M. Preparation of pyrimidine derivatives as Polo-like kinases inhibitors for treatment of cancers (World Patent WO 2004074244, 2004).
Barr, F.A., Sillje, H.H. & Nigg, E.A. Polo-like kinases and the orchestration of cell division. Nat. Rev. Mol. Cell Biol. 5, 429–440 (2004).
Takai, N., Hamanaka, R., Yoshimatsu, J. & Miyakawa, I. Polo-like kinases (Plks) and cancer. Oncogene 24, 287–291 (2005).
Compton, D.A. Spindle assembly in animal cells. Annu. Rev. Biochem. 69, 95–114 (2000).
Sharp, D.J., Rogers, G.C. & Scholey, J.M. Microtubule motors in mitosis. Nature 407, 41–47 (2000).
McInnes, C., Mezna, M. & Fischer, P.M. Progress in the discovery of polo-like kinase inhibitors. Curr. Top. Med. Chem. 5, 181–197 (2005).
Gumireddy, K. et al. ON01910, a non-ATP-competitive small molecule inhibitor of Plk1, is a potent anticancer agent. Cancer Cell 7, 275–286 (2005).
McInnes, C., Meades, C., Mezna, M. & Fischer, P. Benzthiazole-3 oxides useful for the treatment of proliferative disorders (World Patent WO 2004067000, 2004).
van Vugt, M.A. et al. Polo-like kinase-1 is required for bipolar spindle formation but is dispensable for anaphase promoting complex/Cdc20 activation and initiation of cytokinesis. J. Biol. Chem. 279, 36841–36854 (2004).
Sumara, I. et al. Roles of polo-like kinase 1 in the assembly of functional mitotic spindles. Curr. Biol. 14, 1712–1722 (2004).
Khodjakov, A., Copenagle, L., Gordon, M.B., Compton, D.A. & Kapoor, T.M. Minus-end capture of preformed kinetochore fibers contributes to spindle morphogenesis. J. Cell Biol. 160, 671–683 (2003).
Toyoshima-Morimoto, F., Taniguchi, E. & Nishida, E. Plk1 promotes nuclear translocation of human Cdc25C during prophase. EMBO Rep. 3, 341–348 (2002).
Nigg, E.A. Mitotic kinases as regulators of cell division and its checkpoints. Nat. Rev. Mol. Cell Biol. 2, 21–32 (2001).
Hauf, S. et al. The small molecule Hesperadin reveals a role for Aurora B in correcting kinetochore-microtubule attachment and in maintaining the spindle assembly checkpoint. J. Cell Biol. 161, 281–294 (2003).
Niiya, F., Xie, X., Lee, K.S., Inoue, H. & Miki, T. Inhibition of cyclin-dependent kinase 1 induces cytokinesis without chromosome segregation in an ECT2 and MgcRacGAP-dependent manner. J. Biol. Chem. 280, 36502–36509 (2005).
Lampson, M.A., Renduchitala, K., Khodjakov, A. & Kapoor, T.M. Correcting improper chromosome-spindle attachments during cell division. Nat. Cell Biol. 6, 232–237 (2004).
Dogterom, M., Kerssemakers, J.W., Romet-Lemonne, G. & Janson, M.E. Force generation by dynamic microtubules. Curr. Opin. Cell Biol. 17, 67–74 (2005).
Li, X. & Nicklas, R.B. Mitotic forces control a cell-cycle checkpoint. Nature 373, 630–632 (1995).
Waters, J.C., Mitchison, T.J., Rieder, C.L. & Salmon, E.D. The kinetochore microtubule minus-end disassembly associated with poleward flux produces a force that can do work. Mol. Biol. Cell 7, 1547–1558 (1996).
Gergely, F., Draviam, V.M. & Raff, J.W. The ch-TOG/XMAP215 protein is essential for spindle pole organization in human somatic cells. Genes Dev. 17, 336–341 (2003).
Joseph, J., Liu, S.T., Jablonski, S.A., Yen, T.J. & Dasso, M. The RanGAP1-RanBP2 complex is essential for microtubule-kinetochore interactions in vivo. Curr. Biol. 14, 611–617 (2004).
Garrett, S., Auer, K., Compton, D.A. & Kapoor, T.M. hTPX2 is required for normal spindle morphology and centrosome integrity during vertebrate cell division. Curr. Biol. 12, 2055–2059 (2002).
Gruber, J., Harborth, J., Schnabel, J., Weber, K. & Hatzfeld, M. The mitotic-spindle-associated protein astrin is essential for progression through mitosis. J. Cell Sci. 115, 4053–4059 (2002).
Adams, R.R., Maiato, H., Earnshaw, W.C. & Carmena, M. Essential roles of Drosophila inner centromere protein (INCENP) and aurora B in histone H3 phosphorylation, metaphase chromosome alignment, kinetochore disjunction, and chromosome segregation. J. Cell Biol. 153, 865–880 (2001).
Holt, S.V. et al. Silencing Cenp-F weakens centromeric cohesion, prevents chromosome alignment and activates the spindle checkpoint. J. Cell Sci. 118, 4889–4900 (2005).
Zhu, C. et al. Functional analysis of human microtubule-based motor proteins, the kinesins and dyneins, in mitosis/cytokinesis using RNA interference. Mol. Biol. Cell 16, 3187–3199 (2005).
Dai, J., Sultan, S., Taylor, S.S. & Higgins, J.M. The kinase haspin is required for mitotic histone H3 Thr 3 phosphorylation and normal metaphase chromosome alignment. Genes Dev. 19, 472–488 (2005).
McEwen, B.F., Heagle, A.B., Cassels, G.O., Buttle, K.F. & Rieder, C.L. Kinetochore fiber maturation in PtK1 cells and its implications for the mechanisms of chromosome congression and anaphase onset. J. Cell Biol. 137, 1567–1580 (1997).
Knight, Z.A. & Shokat, K.M. Features of selective kinase inhibitors. Chem. Biol. 12, 621–637 (2005).
Tanaka, M. et al. An unbiased cell morphology-based screen for new, biologically active small molecules. PLoS Biol. 3, e128 (2005).
Qian, Y.W., Erikson, E., Li, C. & Maller, J.L. Activated polo-like kinase Plx1 is required at multiple points during mitosis in Xenopus laevis. Mol. Cell. Biol. 18, 4262–4271 (1998).
Acknowledgements
We thank C. Karan and acknowledge the use of the Rockefeller University High Throughput Screening Resource Center. This work was supported by US National Institutes of Health grant GM71772 (to T.M.K.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Fig. 1
Composition of the diaminopyrimidine library. (PDF 194 kb)
Supplementary Fig. 2
Overview of cell-based screening results. (PDF 257 kb)
Supplementary Fig. 3
Mitotic phenotypes induced by a subset of DAPs. (PDF 442 kb)
Supplementary Fig. 4
Monopolar mitotic spindles induced by two diaminopyrimidines do not result from changes in centrosome number or Eg5 inhibition. (PDF 908 kb)
Supplementary Fig. 5
BTO-1 inhibits Polo-like kinase 1 in vitro. (PDF 93 kb)
Supplementary Fig. 6
Additional images for comparing Plk1-knock-down and treatment with chemical inhibitors. (PDF 129 kb)
Supplementary Fig. 7
Quantitation of monopolar spindles and mitotic indices in DAP-81 and BTO-1 treated PTKαT cells. (PDF 97 kb)
Rights and permissions
About this article
Cite this article
Peters, U., Cherian, J., Kim, J. et al. Probing cell-division phenotype space and Polo-like kinase function using small molecules. Nat Chem Biol 2, 618–626 (2006). https://doi.org/10.1038/nchembio826
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nchembio826
This article is cited by
-
Identification of inhibitors of the polo-box domain of polo-like kinase 1 from natural and semisynthetic compounds
Investigational New Drugs (2020)
-
Chemical dissection of the cell cycle: probes for cell biology and anti-cancer drug development
Cell Death & Disease (2014)
-
Plk1-Targeted Small Molecule Inhibitors: Molecular Basis for Their Potency and Specificity
Molecules and Cells (2011)
-
Shared and separate functions of polo-like kinases and aurora kinases in cancer
Nature Reviews Cancer (2010)
-
Multifaceted polo-like kinases: drug targets and antitargets for cancer therapy
Nature Reviews Drug Discovery (2010)