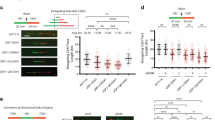
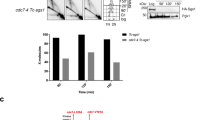
Abstract
In all eukaryotes, the ligation of newly synthesized DNA, also known as Okazaki fragments, is catalysed by DNA ligase I (ref. 1). An individual with a DNA ligase I deficiency exhibits growth retardation, sunlight sensitivity and severe immunosuppression2, probably due to accumulation of DNA damage. Surprisingly, not much is known about the DNA damage response (DDR) in DNA ligase I-deficient cells. As DNA replication and DDR pathways are highly conserved in eukaryotes, we used Saccharomyces cerevisiae as a model system to address this issue. We uncovered a new pathway, which facilitates ubiquitylation at Lys 107 of proliferating cell nuclear antigen (PCNA). Unlike ubiquitylation at Lys 164 of PCNA in response to UV irradiation, which triggers translesion synthesis3, modification of Lys 107 is not dependent on the ubiquitin conjugating enzyme (E2) Rad6 (ref. 4) nor the ubiquitin ligase (E3) Rad18 (ref. 5), but requires the E2 variant Mms2 (ref. 6) in conjunction with Ubc4 (ref. 7) and the E3 Rad5 (Refs 8, 9). Surprisingly, DNA ligase I-deficient S. cerevisiae cdc9-1 cells that carry a PCNAK107R mutation are inviable, because they cannot activate a robust DDR. Furthermore, we show that ubiquitylation of PCNA in response to DNA ligase I deficiency is conserved in humans, yet the lysine residue that is modified remains to be determined. We propose that PCNA ubiquitylation provides a 'DNA damage code' that allows cells to categorize different types of defects that arise during DNA replication.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout





Similar content being viewed by others
References
Ellenberger, T. & Tomkinson, A. E. Eukaryotic DNA ligases: structural and functional insights. Annu. Rev. Biochem 77, 313–338 (2008).
Webster, A. D., Barnes, D. E., Arlett, C. F., Lehmann, A. R. & Lindahl, T. Growth retardation and immunodeficiency in a patient with mutations in the DNA ligase I gene. Lancet 339, 1508–1509 (1992).
Hoege, C., Pfander, B., Moldovan, G. L., Pyrowolakis, G. & Jentsch, S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419, 135–141 (2002).
Jentsch, S., McGrath, J. P. & Varshavsky, A. The yeast DNA repair gene RAD6 encodes a ubiquitin-conjugating enzyme. Nature 329, 131–134 (1987).
Bailly, V., Lauder, S., Prakash, S. & Prakash, L. Yeast DNA repair proteins Rad6 and Rad18 form a heterodimer that has ubiquitin conjugating, DNA binding, and ATP hydrolytic activities. J. Biol. Chem. 272, 23360–23365 (1997).
Broomfield, S., Chow, B. L. & Xiao, W. MMS2, encoding a ubiquitin-conjugating-enzyme-like protein, is a member of the yeast error-free postreplication repair pathway. Proc. Natl Acad. Sci. USA 95, 5678–5683 (1998).
Seufert, W. & Jentsch, S. Ubiquitin-conjugating enzymes UBC4 and UBC5 mediate selective degradation of short-lived and abnormal proteins. EMBO J. 9, 543–550 (1990).
Ulrich, H. D. & Jentsch, S. Two RING finger proteins mediate cooperation between ubiquitin-conjugating enzymes in DNA repair. EMBO J. 19, 3388–3397 (2000).
Torres-Ramos, C. A., Prakash, S. & Prakash, L. Requirement of RAD5 and MMS2 for postreplication repair of UV-damaged DNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 22, 2419–2426 (2002).
Schiestl, R. H., Reynolds, P., Prakash, S. & Prakash, L. Cloning and sequence analysis of the Saccharomyces cerevisiae RAD9 gene and further evidence that its product is required for cell cycle arrest induced by DNA damage. Mol. Cell. Biol. 9, 1882–1896 (1989).
Weinert, T. A. & Hartwell, L. H. Cell cycle arrest of cdc mutants and specificity of the RAD9 checkpoint. Genetics 134, 63–80 (1993).
Johnston, L. H. & Nasmyth, K. A. Saccharomyces cerevisiae cell cycle mutant cdc9 is defective in DNA ligase. Nature 274, 891–893 (1978).
Dohmen, R. J., Wu, P. & Varshavsky, A. Heat-inducible degron: a method for constructing temperature-sensitive mutants. Science 263, 1273–1276 (1994).
Bielinsky, A. K. & Gerbi, S. A. Chromosomal ARS1 has a single leading strand start site. Mol. Cell 3, 477–486 (1999).
Ireland, M. J., Reinke, S. S. & Livingston, D. M. The impact of lagging strand replication mutations on the stability of CAG repeat tracts in yeast. Genetics 155, 1657–1665 (2000).
Osborn, A. J. & Elledge, S. J. Mrc1 is a replication fork component whose phosphorylation in response to DNA replication stress activates Rad53. Genes Dev. 17, 1755–1767 (2003).
Sanchez, Y. et al. Regulation of RAD53 by the ATM-like kinases MEC1 and TEL1 in yeast cell cycle checkpoint pathways. Science 271, 357–360 (1996).
Moldovan, G. L., Pfander, B. & Jentsch, S. PCNA, the maestro of the replication fork. Cell 129, 665–679 (2007).
Stelter, P. & Ulrich, H. D. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature 425, 188–191 (2003).
Haracska, L., Torres-Ramos, C. A., Johnson, R. E., Prakash, S. & Prakash, L. Opposing effects of ubiquitin conjugation and SUMO modification of PCNA on replicational bypass of DNA lesions in Saccharomyces cerevisiae. Mol. Cell. Biol. 24, 4267–4274 (2004).
Hofmann, R. M. & Pickart, C. M. Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell 96, 645–653 (1999).
Zhang, Z., Shibahara, K. & Stillman, B. PCNA connects DNA replication to epigenetic inheritance in yeast. Nature 408, 221–225 (2000).
Ellison, M. J. & Hochstrasser, M. Epitope-tagged ubiquitin. A new probe for analyzing ubiquitin function. J. Biol. Chem. 266, 21150–21157 (1991).
Das-Bradoo, S., Ricke, R. M. & Bielinsky, A. K. Interaction between PCNA and diubiquitinated Mcm10 is essential for cell growth in budding yeast. Mol. Cell. Biol. 26, 4806–4817 (2006).
van der Kemp, P. A., Padula, M. D., Burguiere-Slezak, G., Ulrich, H. D. & Boiteux, S. PCNA monoubiquitylation and DNA polymerase-eta ubiquitin-binding domain are required to prevent 8-oxoguanine-induced mutagenesis in Saccharomyces cerevisiae. Nucleic Acids Res. 37, 2549–2559 (2009).
Wang, M. & Pickart, C. M. Different HECT domain ubiquitin ligases employ distinct mechanisms of polyubiquitin chain synthesis. EMBO J. 24, 4324–4333 (2005).
Pettersen, E. F. et al. UCSF Chimera-a visualization system for exploratory research and analysis. J. Comput. Chem. 25, 1605–1612 (2004).
Frampton, J. et al. Postreplication repair and PCNA modification in Schizosaccharomyces pombe. Mol. Biol. Cell 17, 2976–2985 (2006).
Kannouche, P. L., Wing, J. & Lehmann, A. R. Interaction of human DNA polymerase eta with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol. Cell 14, 491–500.
Kao, H. I., Veeraraghavan, J., Polaczek, P., Campbell, J. L. & Bambara, R. A. On the roles of Saccharomyces cerevisiae Dna2p and Flap endonuclease 1 in Okazaki fragment processing. J. Biol. Chem. 279, 15014–15024 (2004).
Labib, K., Tercero, J. A. & Diffley, J. F. Uninterrupted MCM2–7 function required for DNA replication fork progression. Science 288, 1643–1647 (2000).
Lorenz, M. C. et al. Gene disruption with PCR products in Saccharomyces cerevisiae. Gene 158, 113–117 (1995).
Wen, W., Meinkoth, J. L., Tsien, R. Y. & Taylor, S. S. Identification of a signal for rapid export of proteins from the nucleus. Cell 82, 463–473 (1995).
Ricke, R. M. & Bielinsky, A. K. Mcm10 regulates the stability and chromatin association of DNA polymerase-α. Mol. Cell 16, 173–185 (2004).
Ricke, R. M. & Bielinsky, A. K. A conserved Hsp10-like domain in Mcm10 is required to stabilize the catalytic subunit of DNA polymerase-α in budding yeast. J. Biol. Chem. 281, 18414–18425 (2006).
Diffley, J. F. Interdependent nuclear accumulation of budding yeast Cdt1 and Mcm2–7 during G1 phase. Nature Cell Biol. 4, 198–207 (2002).
Silva, J. M. et al. Second-generation shRNA libraries covering the mouse and human genomes. Nature Genetics 37, 1281–1288 (2005).
Hannon, G. J. & Conklin, D. S. RNA interference by short hairpin RNAs expressed in vertebrate cells. Methods Mol. Biol. 257, 255–266 (2004).
Motegi, A. et al. Polyubiquitination of proliferating cell nuclear antigen by HLTF and SHPRH prevents genomic instability from stalled replication fork. Proc. Natl Acad. Sci. USA 26, 12411–12416 (2008).
Acknowledgements
We thank D. J. Clarke, J. F. X. Diffley, D. Durocher, S. J. Elledge, M. Hochstrasser, S. Jentsch, D. Koepp and D. M. Livingston for strains and plasmids and J. F. X. Diffley, A. E. Tomkinson and Z. Zhang for antibodies against Rad53, Cdc9 and PCNA. We acknowledge the assistance of the Flow Cytometry Core Facility at the University of Minnesota. Molecular graphics images were produced using the UCSF Chimera package from the Resource for Biocomputing, Visualization, and Informatics at the University of California, San Francisco (supported by NIH P41 RR-01081). J.C.H. was supported by NIH training grant CA009138. This work was supported by a Grant-in-Aid from the University of Minnesota and NIH grant (GM074917) to A.K.B. who is a Scholar of the Leukemia and Lymphoma Society.
Author information
Authors and Affiliations
Contributions
S.D.-B. and H.D.N. conducted all PCNA ubiquitylation experiments in yeast, helped with data analysis and helped write the manuscript. J.L.W. conducted all experiments with human cells, helped with data analysis and helped write the manuscript. R.M.R conducted cell-cycle arrest and RIP mapping experiments, and helped with data analysis. J.C.H. helped with the construction of yeast mutants. A.-K.B. planned and supervised the project, and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Information (PDF 1867 kb)
Rights and permissions
About this article
Cite this article
Das-Bradoo, S., Nguyen, H., Wood, J. et al. Defects in DNA ligase I trigger PCNA ubiquitylation at Lys 107. Nat Cell Biol 12, 74–79 (2010). https://doi.org/10.1038/ncb2007
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/ncb2007
This article is cited by
-
Sumo and the cellular stress response
Cell Division (2015)
-
Molecular architecture of the Ub-PCNA/Pol η complex bound to DNA
Scientific Reports (2015)
-
In vivo and in silico analysis of PCNA ubiquitylation in the activation of the Post Replication Repair pathway in S. cerevisiae
BMC Systems Biology (2013)
-
Regulation of PCNA–protein interactions for genome stability
Nature Reviews Molecular Cell Biology (2013)
-
Partial complementation of a DNA ligase I deficiency by DNA ligase III and its impact on cell survival and telomere stability in mammalian cells
Cellular and Molecular Life Sciences (2012)