Abstract
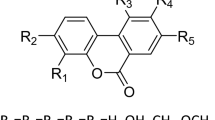
Cyclic nucleotide phosphodiesterases (PDEs) comprise a large family of enzymes that regulate a variety of cellular processes. We describe a family of potent PDE4 inhibitors discovered using an efficient method for scaffold-based drug design. This method involves an iterative approach starting with low-affinity screening of compounds followed by high-throughput cocrystallography to reveal the molecular basis underlying the activity of the newly identified compounds. Through detailed structural analysis of the interaction of the initially discovered pyrazole carboxylic ester scaffold with PDE4D using X-ray crystallography, we identified three sites of chemical substitution and designed small selective libraries of scaffold derivatives with modifications at these sites. A 4,000-fold increase in the potency of this PDE4 inhibitor was achieved after only two rounds of chemical synthesis and the structural analysis of seven pyrazole derivatives bound to PDE4B or PDE4D, revealing the robustness of this approach for identifying new inhibitors that can be further developed into drug candidates.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Lipinski, C.A., Lombardo, F., Dominy, B.W. & Feeney, P.J. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Rev. 23, 3–25 (1997).
Erlanson, D.A., McDowell, R.S. & O'Brien, T. Fragment-based drug discovery. J. Med. Chem. 47, 3463–3482 (2004).
Rees, D.C., Congreve, M., Murray, C.W. & Carr, R. Fragment-based lead discovery. Nat. Rev. Drug Discov. 3, 660–672 (2004).
Shuker, S.B., Hajduk, P.J., Meadows, R.P. & Fesik, S.W. Discovering high-affinity ligands for proteins: SAR by NMR. Science 274, 1531–1534 (1996).
Nienaber, V.L. et al. Discovering novel ligands for macromolecules using X-ray crystallographic screening. Nat. Biotechnol. 18, 1105–1108 (2000).
Erlanson, D.A., Wells, J.A. & Braisted, A.C. Tethering: fragment-based drug discovery. Annu. Rev. Biophys. Biomol. Struct. 33, 199–223 (2004).
Houslay, M.D. Adaptation in cyclic AMP signalling processes: a central role for cyclic AMP phosphodiesterases. Semin. Cell Dev. Biol. 9, 161–167 (1998).
Beavo, J.A. Cyclic nucleotide phosphodiesterases: functional implications of multiple isoforms. Physiol. Rev. 75, 725–748 (1995).
Conti, M. & Jin, S.L. The molecular biology of cyclic nucleotide phosphodiesterases. Prog. Nucleic Acid Res. Mol. Biol. 63, 1–38 (1999).
Francis, S.H., Turko, I.V. & Corbin, J.D. Cyclic nucleotide phosphodiesterases: relating structure and function. Prog. Nucleic Acid Res. Mol. Biol. 65, 1–52 (2001).
Corbin, J.D. & Francis, S.H. Pharmacology of phosphodiesterase-5 inhibitors. Int. J. Clin. Pract. 56, 453–459 (2002).
Rotella, D.P. Phosphodiesterase 5 inhibitors: current status and potential applications. Nat. Rev. Drug Discov. 1, 674–682 (2002).
Souness, J.E., Aldous, D. & Sargent, C. Immunosuppressive and anti-inflammatory effects of cyclic AMP phosphodiesterase (PDE) type 4 inhibitors. Immunopharmacology 47, 127–162 (2000).
Mehats, C., Andersen, C.B., Filopanti, M., Jin, S.L. & Conti, M. Cyclic nucleotide phosphodiesterases and their role in endocrine cell signaling. Trends Endocrinol. Metab. 13, 29–35 (2002).
Beavo, J.A. & Brunton, L.L. Cyclic nucleotide research–still expanding after half a century. Nat. Rev. Mol. Cell Biol. 3, 710–718 (2002).
Conti, M. Phosphodiesterases and cyclic nucleotide signaling in endocrine cells. Mol. Endocrinol. 14, 1317–1327 (2000).
Conti, M. et al. Cyclic AMP-specific PDE4 phosphodiesterases as critical components of cyclic AMP signaling. J. Biol. Chem. 278, 5493–5496 (2003).
Houslay, M.D. & Adams, D.R. PDE4 cAMP phosphodiesterases: modular enzymes that orchestrate signalling cross-talk, desensitization and compartmentalization. Biochem. J. 370, 1–18 (2003).
Jin, S.L. & Conti, M. Induction of the cyclic nucleotide phosphodiesterase PDE4B is essential for LPS-activated TNF-alpha responses. Proc. Natl. Acad. Sci. USA 99, 7628–7633 (2002).
Bardelle, C. et al. Phosphodiesterase 4 conformers: preparation of recombinant enzymes and assay for inhibitors. Anal. Biochem. 275, 148–155 (1999).
Card, G.L. et al. Structural basis for the activity of drugs that inhibit phosphodiesterases. Structure 12, 2233–2247 (2004).
Srinivasan, J., Cheatham, T.E., Cieplak, P., Kollman, P.A. & Case, D.A. Continuum solvent studies of the stability of DNA, RNA, and phosphoramidate-DNA helices. J. Am. Chem. Soc. 120, 9401–9409 (1998).
Case, D.A. et al. AMBER 7 (University of California, San Francisco, 2002).
Zhang, K.Y.J. et al. A glutamine switch mechanism for nucleotide selectivity by phosphodiesterases. Mol. Cell 15, 279–286 (2004).
Richter, W. 1,3-Cyclopentanedione and isomeric enol-lactones. IV. Some reactions of diacetic acid ethyl ester. Helv. Chim. Acta 35, 478–485 (1952).
Cornell, W.D. et al. A second generation force field for the simulation of proteins, nucleic acid, and organic molecules. J. Am. Chem. Soc. 117, 5179–5197 (1995).
Stewart, J.J.P. MOPAC Manual (United States Air Force Academy, Colorado Springs, CO, 1985).
Leslie, A.G. Integration of macromolecular diffraction data. Acta Crystallogr. D55, 1696–1702 (1999).
Evans, P.R. Data Reduction. Proceedings of CCP4 Study Weekend, On Data Collection and Processing, Daresbury, Warrington, UK, 114–122 (1993).
Holton, J. & Alber, T. From the cover: automated protein crystal structure determination using ELVES. Proc. Natl. Acad. Sci. USA 101, 1537–1542 (2004).
Xu, R.X. et al. Atomic structure of PDE4: insights into phosphodiesterase mechanism and specificity. Science 288, 1822–1825 (2000).
Huai, Q. et al. Three-dimensional structures of PDE4D in complex with roliprams and implication on inhibitor selectivity. Structure 11, 865–873 (2003).
Brünger, A.T. et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D54, 905–921 (1998).
Murshudov, G.N., Vagin, A.A., Lebedev, A., Wilson, K.S. & Dodson, E.J. Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr. D55, 247–255 (1999).
Jones, T.A., Zou, J.Y., Cowan, S.W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A47, 110–119 (1991).
Acknowledgements
We thank Peter Hirth for his insight and inspiration on the scaffold-based drug design paradigm. We thank Axel T. Brunger for critical reading of this manuscript and for discussions. We also thank our colleagues at Plexxikon for help and discussions. We acknowledge Jim Arnold, Fernando Martin and Richard Fronko for the scaffold library design and construction. We thank Ben Powell, Catherine Luu and Hoa Nguyen for protein purification. We thank Jaina Thompson for compound preparation for screening and cocrystallization. We thank Heike Krupka, Abhinav Kumar and Weiru Wang for assisting in crystal mounting and data collection, and Abhinav Kumar for assisting in structure determination. Diffraction data were collected at the Advanced Light Source and the Stanford Synchrotron Radiation Laboratory, which are supported by the US Department of Energy, Office of Basic Energy Sciences under contract DE-AC03-76SF00098 and DE-AC03-76SF00515, respectively. J.S. is supported by National Institutes of Health grant 1RO1-AR051448-01 and funds from the Ludwig Institute for Cancer Research.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
All the authors except S.-H.K. and J.S. are employees of Plexxikon. S.-H.K. and J.S. are cofounders of the company and shareholders of Plexxikon.
Supplementary information
Supplementary Table 1
Biochemical inhibition data of the pyrazoles against a full panel of PDEs. (PDF 213 kb)
Rights and permissions
About this article
Cite this article
Card, G., Blasdel, L., England, B. et al. A family of phosphodiesterase inhibitors discovered by cocrystallography and scaffold-based drug design. Nat Biotechnol 23, 201–207 (2005). https://doi.org/10.1038/nbt1059
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt1059
This article is cited by
-
DrugEx v3: scaffold-constrained drug design with graph transformer-based reinforcement learning
Journal of Cheminformatics (2023)
-
Vemurafenib: the first drug approved for BRAF-mutant cancer
Nature Reviews Drug Discovery (2012)
-
Hit clustering can improve virtual fragment screening: CDK2 and PARP1 case studies
Journal of Molecular Modeling (2012)
-
Computational fragment-based screening using RosettaLigand: the SAMPL3 challenge
Journal of Computer-Aided Molecular Design (2012)