Abstract
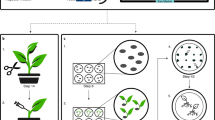
It is generally thought that transformation of plant cells using Agrobacterium tumefaciens occurs at a very low frequency. Therefore, selection marker genes are used to identify the rare plants that have taken up foreign DNA. Genes encoding antibiotic and herbicide resistance are widely used for this purpose in plant transformation1,2. Over the past several years, consumer and environmental groups have expressed concern about the use of antibiotic- and herbicide-resistance genes from an ecological and food safety perspective. Although no scientific basis has been determined for these concerns, generating marker-free plants would certainly contribute to the public acceptance of transgenic crops. Several methods have been reported to create marker gene–free transformed plants, for example co-transformation, transposable elements, site-specific recombination, or intrachromosomal recombination3,4,5,6,7,8,9. Not only are most of these systems time-consuming and inefficient, but they are also employed on the assumption that isolation of transformants without a selective marker gene is not feasible10. Here we present a method that permits the identification of transgenic plants without the use of selectable markers. This strategy relies on the transformation of tissue explants or cells with a virulent A. tumefaciens strain and selection of transformed cells or shoots after PCR analysis. Incubation of potato explants with A. tumefaciens strain AGL0 resulted in transformed shoots at an efficiency of 1–5% of the harvested shoots, depending on the potato genotype used. Because this system does not require genetic segregation or site-specific DNA-deletion systems to remove marker genes, it may provide a reliable and efficient tool for generating transgenic plants for commercial use, especially in vegetatively propagated species like potato and cassava.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 12 print issues and online access
$209.00 per year
only $17.42 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Bevan, M.W., Flavell, R.B. & Chilton, M.D. A chimaeric antibiotic resistance gene as a selection marker for plant cell transformation. Nature 304, 184–187 (1983).
Shah, D.M. et al. Engineering herbicide tolerance in transgenic plants. Science 233, 478–481 (1986).
Komari, T., Hiei, Y., Saito, Y., Murai, N. & Kumashiro, T. Vectors carrying two separate T-DNAs for co-transformation of higher plants mediated by Agrobacterium tumefaciens and segregation of transformants free from selection markers. Plant J. 10, 165–174 (1996).
Gleave, A.P., Mitra, D.S., Mudge, S.R. & Morris, B.A.M. Selectable marker-free transgenic plants without sexual crossing: transient expression of cre recombinase and use of a conditional lethal dominant gene. Plant Mol. Biol. 40, 223–235 (1999).
Goldbrough, A.P., Lastrella, C.N. & Yoder, J.I. Transposition mediated re-positioning and subsequent elimination of marker genes from transgenic tomato. Bio/technology 11, 1286–1292 (1993).
Ebinuma, H., Sugita, K., Matsunaga, E. & Yamakado, M. Selection of marker-free transgenic plants using the isopentenyl transferase gene. Proc. Natl. Acad. Sci. USA 94, 2117–2121 (1997).
Dale, E.C. & Ow, D.W. Gene transfer with subsequent removal of the selection gene from the host genome. Proc. Natl. Acad. Sci. USA 88, 10558–10562 (1991).
Zuo, J., Nui, Q.-W., Moller, S.G. & Chua, N.-H. Chemical-regulated, site-specific DNA excision in transgenic plants. Nat. Biotechnol. 19, 157–161 (2001).
Zubko, E., Scutt, C. & Meyer, P. Intrachromosomal recombination between attP regions as a tool to remove selectable marker genes from tobacco transgenes. Nat. Biotechnol. 18, 442–445 (2000).
Ow, D.W. The right chemistry for marker gene removal? Nat. Biotechnol. 19, 115–116 (2001).
Hood, E.E., Helmer, G.L., Fraley, R.T. & Shilton, M.D. The hypervirulence of Agrobacterium tumefaciens A281 is encoded in a region of pTiBo542 outside of the T-DNA. J. Bacteriol. 168, 1291–1304 (1986).
Murashige, T. & Skoog, F. A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant 15, 473–497 (1962).
Heeres, P., Schippers-Rozenboom, M., Jacobsen, E. & Visser, R.G.F. Transformation of a large number of potato varieties: genotype dependent variation in efficiency and somaclonal variability. Euphytica 124, 13–22 (2002).
Martineau, B., Voelker, T.A. & Sanders, R.A. On defining T-DNA. Plant Cell 6, 1032–1033 (1994).
Kononov, M.E., Bassuner, B. & Gelvin, S.B. Integration of T-DNA binary vector “backbone” sequences into the tobacco genome: evidence for multiple complex patterns of integration. Plant J. 11, 945–957 (1997).
Wolters, A.-M.A., Trindade, L.M., Jacobsen, E. & Visser, R.G.F. Fluorescence in situ hybridization on extended DNA fibers as a tool to analyse complex T-DNA loci in potato. Plant J. 13, 837–847 (1998).
Lazo, G.R., Stein, P.A. & Ludwig, R.A. DNA transformation-competent Arabidopsis genomic library in Agrobacterium. Bio/Technology 9, 963–967 (1991).
Raemakers, C.J.J.M., Sofiari, E., Taylor, N., Henshaw, G., Jacobsen, E. & Visser, R.G.F. Production of transgenic cassava (Manihot esculenta Crantz) plants by particle bombardment using luciferase activity as selection marker. Mol. Breeding 2, 339–349 (1996).
Kuipers, A.G.J., Soppe, W.J.J., Jacobsen, E. & Visser, R.G.F. Factors affecting the inhibition by antisense RNA of granule-bound starch synthase gene expression in potato. Mol. Gen. Genet. 246, 745–755 (1995).
Hoekema, A., Hirsch, P.R., Hooykaas, P.J.J. & Schilperoort, R.A. A binary plant vector strategy based on separation of vir- and T-region of the Agrobacterium tumefaciens Ti-plasmid. Nature 303, 179–180 (1983).
Visser, R.G.F. Regeneration and transformation of potato by Agrobacterium tumefaciens. in Plant Tissue Culture Manual (ed. Lindsey, K.) Sec. B5, 1–9 (Kluwer Academic Publishers, Dordrecht, Boston, London, 1991).
Rogers, S.O. & Bendich, A.J. Extraction of DNA from plant tissues. in Plant Molecular Biology Manual A6 (eds. Gelvin, S.B. & Schilperoort, R.A.) Sec. A6, 1–10 (Kluwer Academic Publishers, Dordrecht, 1988).
Schreuder, M.M., Raemakers, C.J.J.M., Jacobsen, E. & Visser, R.G.F. Efficient production of transgenic plants by Agrobacterium-mediated transformation of cassava (Manihot esculenta Crantz). Euphytica 120, 35–42 (2001).
Acknowledgements
The authors wish to thank SENTER for financial support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
About this article
Cite this article
de Vetten, N., Wolters, AM., Raemakers, K. et al. A transformation method for obtaining marker-free plants of a cross-pollinating and vegetatively propagated crop. Nat Biotechnol 21, 439–442 (2003). https://doi.org/10.1038/nbt801
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nbt801