Abstract
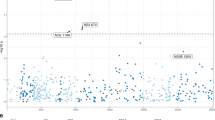
Chronic infection with hepatitis C virus (HCV) affects 170 million people worldwide and is the leading cause of cirrhosis in North America1. Although the recommended treatment for chronic infection involves a 48-week course of peginterferon-α-2b (PegIFN-α-2b) or -α-2a (PegIFN-α-2a) combined with ribavirin (RBV), it is well known that many patients will not be cured by treatment, and that patients of European ancestry have a significantly higher probability of being cured than patients of African ancestry. In addition to limited efficacy, treatment is often poorly tolerated because of side effects that prevent some patients from completing therapy. For these reasons, identification of the determinants of response to treatment is a high priority. Here we report that a genetic polymorphism near the IL28B gene, encoding interferon-λ-3 (IFN-λ-3), is associated with an approximately twofold change in response to treatment, both among patients of European ancestry (P = 1.06 × 10-25) and African-Americans (P = 2.06 × 10-3). Because the genotype leading to better response is in substantially greater frequency in European than African populations, this genetic polymorphism also explains approximately half of the difference in response rates between African-Americans and patients of European ancestry.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
Change history
17 September 2009
The x-axis label in Figure 3 was corrected on 17 September 2009.
References
World Health Organization. Hepatitis C. 〈http://www.who.int/mediacentre/factsheets/fs164/en/〉 (2009)
McHutchison, J. G. et al. Peginterferon alfa-2b or alfa-2a with ribavirin for treatment of hepatitis C infection. N. Engl. J. Med. 361, 580–593 (2009)
Muir, A. J., Bornstein, J. D. & Killenberg, P. G. Peginterferon alfa-2b and ribavirin for the treatment of chronic hepatitis C in blacks and non-Hispanic whites. N. Engl. J. Med. 350, 2265–2271 (2004)
Ghany, M. G., Strader, D. B., Thomas, D. L. & Seeff, L. B. Diagnosis, management, and treatment of hepatitis C: an update. Hepatology 49, 1335–1374 (2009)
Shiffman, M. L. et al. Peginterferon alfa-2a and ribavirin in patients with chronic hepatitis C who have failed prior treatment. Gastroenterology 126, 1015–1023; discussion 947 (2004)
Yan, K. K. et al. Treatment responses in Asians and Caucasians with chronic hepatitis C infection. World J. Gastroenterol. 14, 3416–3420 (2008)
Liu, C. H. et al. Pegylated interferon-alpha-2a plus ribavirin for treatment-naive Asian patients with hepatitis C virus genotype 1 infection: a multicenter, randomized controlled trial. Clin. Infect. Dis. 47, 1260–1269 (2008)
Wilson, J. F. et al. Population genetic structure of variable drug response. Nature Genet. 29, 265–269 (2001)
Sarasin-Filipowicz, M. et al. Interferon signaling and treatment outcome in chronic hepatitis C. Proc. Natl Acad. Sci. USA 105, 7034–7039 (2008)
McHutchison, J. G. et al. Telaprevir with peginterferon and ribavirin for chronic HCV genotype 1 infection. N. Engl. J. Med. 360, 1827–1838 (2009)
Robek, M. D., Boyd, B. S. & Chisari, F. V. Lambda interferon inhibits hepatitis B and C virus replication. J. Virol. 79, 3851–3854 (2005)
Shiffman, M. L. et al. PEG-IFN-λ: antiviral activity and safety profile in a 4-week phase 1b study in relapsed genotype 1 hepatitis C infection. J. Hepatol. 50 (suppl. 1), abstr. A643 s237 (2009)
Kotenko, S. V. et al. IFN-λs mediate antiviral protection through a distinct class II cytokine receptor complex. Nature Immunol. 4, 69–77 (2003)
Sheppard, P. et al. IL-28, IL-29 and their class II cytokine receptor IL-28R. Nature Immunol. 4, 63–68 (2003)
Purcell, S. et al. PLINK: a toolset for whole-genome association and population-based linkage analysis. Am. J. Hum. Genet. 81, 559–575 (2007)
Whitlock, M. C. Combining probability from independent tests: the weighted Z-method is superior to Fisher’s approach. J. Evol. Biol. 18, 1368–1373 (2005)
Price, A. L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nature Genet. 38, 904–909 (2006)
The French METAVIR Cooperative Study Group Intraobserver and interobserver variations in liver biopsy interpretation in patients with chronic hepatitis C. Hepatology 20, 15–20 (1994)
Ge, D. et al. WGAViewer: software for genomic annotation of whole genome association studies. Genome Res. 18, 640–643 (2008)
Acknowledgements
We are indebted to the IDEAL principal investigators, the study coordinators, nurses and patients involved in the study. We also recognize E. Gustafson, P. Savino, D. Devlin, S. Noviello, M. Geffner, J. Albrecht and A. C. Need for their contributions to the study. This study was funded by Schering-Plough Research Institute, Kenilworth, New Jersey. A.J.T. received funding support from the National Health and Medical Research Council of Australia and the Gastroenterological Society of Australia.
Author Contributions D.G., J.F., A.J.T. and J.S.S. contributed equally to this work. D.G., J.F. and A.J.T. performed the statistical and bioinformatical analyses. J.F. and A.J.T. defined the clinical phenotypes. K.V.S. performed the genotyping. D.B.G. and D.G. drafted the manuscript. J.S.S., J.G.M. and D.B.G. designed the study. All authors collected and analysed data and contributed to preparing the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Competing Interests: J.G.M. and D.B.G. received research and grant support from Schering-Plough. J.G.M., A.J.M., M.S. and D.B.G. received consulting fees or acted in an advisory capacity for Schering-Plough. J.S.S., P.Q. and A.H.B. are employees of Schering-Plough. D.G., J.F., J.S.S., K.V.S., T.J.U., A.H.B. and D.B.G. are inventors of a patent application based on this finding.
Supplementary information
Supplementary Information
This file contains Supplementary Notes, Supplementary Tables S1-S8, Supplementary Box S1, Supplementary Figure S1 with Legend and Supplementary References. (PDF 440 kb)
Rights and permissions
About this article
Cite this article
Ge, D., Fellay, J., Thompson, A. et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature 461, 399–401 (2009). https://doi.org/10.1038/nature08309
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature08309
This article is cited by
-
The relation between SNPs in the NME1 gene and response to sofosbuvir in Egyptian patients with chronic HCV
Beni-Suef University Journal of Basic and Applied Sciences (2023)
-
Response to antiviral therapy for chronic hepatitis C and risk of hepatocellular carcinoma occurrence in Japan: a systematic review and meta-analysis of observational studies
Scientific Reports (2023)
-
Genetic variants of interleukin 1B and 6 are associated with clinical outcome of surgically treated lumbar degenerative disc disease
BMC Musculoskeletal Disorders (2022)
-
Breakthroughs in hepatitis C research: from discovery to cure
Nature Reviews Gastroenterology & Hepatology (2022)
-
The role of IFNL4 in liver inflammation and progression of fibrosis
Genes & Immunity (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.