Abstract
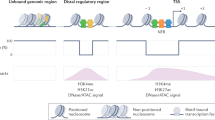
The regulation of gene transcription involves a dynamic balance between packaging regulatory sequences into chromatin and allowing transcriptional regulators access to these sequences. Access is restricted by the nucleosomes, but these can be repositioned or ejected by enzymes known as nucleosome remodellers. In addition, the DNA sequence can impart stiffness or curvature to the DNA, thereby affecting the position of nucleosomes on the DNA, influencing particular promoter 'architectures'. Recent genome-wide studies in yeast suggest that constitutive and regulated genes have architectures that differ in terms of nucleosome position, turnover, remodelling requirements and transcriptional noise.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Li, B., Carey, M. & Workman, J. L. The role of chromatin during transcription. Cell 128, 707–719 (2007).
Narlikar, G. J., Fan, H. Y. & Kingston, R. E. Cooperation between complexes that regulate chromatin structure and transcription. Cell 108, 475–487 (2002).
Becker, P. B. & Horz, W. ATP-dependent nucleosome remodeling. Annu. Rev. Biochem. 71, 247–273 (2002).
Saha, A., Wittmeyer, J. & Cairns, B. R. Chromatin remodelling: the industrial revolution of DNA around histones. Nature Rev. Mol. Cell Biol. 7, 437–447 (2006).
Lee, C. K., Shibata, Y., Rao, B., Strahl, B. D. & Lieb, J. D. Evidence for nucleosome depletion at active regulatory regions genome-wide. Nature Genet. 36, 900–905 (2004).
Lee, W. et al. A high-resolution atlas of nucleosome occupancy in yeast. Nature Genet. 39, 1235–1244 (2007).
Yuan, G. C. et al. Genome-scale identification of nucleosome positions in S. cerevisiae . Science 309, 626–630 (2005). This paper provides the first high-resolution view of nucleosome positioning and reveals a nucleosome-depleted region that overlaps with transcription-factor binding sites.
Barski, A. et al. High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837 (2007).
Bernstein, B. E., Liu, C. L., Humphrey, E. L., Perlstein, E. O. & Schreiber, S. L. Global nucleosome occupancy in yeast. Genome Biol. 5, R62 (2004).
Albert, I. et al. Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature 446, 572–576 (2007).
Mavrich, T. N. et al. Nucleosome organization in the Drosophila genome. Nature 453, 358–362 (2008).
Venters, B. J. & Pugh, B. F. A canonical promoter organization of the transcription machinery and its regulators in the Saccharomyces genome. Genome Res. 19, 360–371 (2009).
Schones, D. E. et al. Dynamic regulation of nucleosome positioning in the human genome. Cell 132, 887–898 (2008).
Segal, E. et al. A genomic code for nucleosome positioning. Nature 442, 772–778 (2006). This influential paper provides computational prediction of nucleosome positions.
Kaplan, N. et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature 458, 362–366 (2009).
Segal, E. & Widom, J. Poly(dA:dT) tracts: major determinants of nucleosome organization. Curr. Opin. Struct. Biol. 19, 65–71 (2009).
Ioshikhes, I. P., Albert, I., Zanton, S. J. & Pugh, B. F. Nucleosome positions predicted through comparative genomics. Nature Genet. 38, 1210–1215 (2006).
Tirosh, I. & Barkai, N. Two strategies for gene regulation by promoter nucleosomes. Genome Res. 18, 1084–1091 (2008). Several concepts in promoter classification developed in this paper are central to my Review.
Field, Y. et al. Distinct modes of regulation by chromatin encoded through nucleosome positioning signals. PLoS Comput. Biol. 4, e1000216 (2008).
Guillemette, B. et al. Variant histone H2A.Z is globally localized to the promoters of inactive yeast genes and regulates nucleosome positioning. PLoS Biol. 3, e384 (2005).
Raisner, R. M. et al. Histone variant H2A.Z marks the 5′ ends of both active and inactive genes in euchromatin. Cell 123, 233–248 (2005).
Zhang, H., Roberts, D. N. & Cairns, B. R. Genome-wide dynamics of Htz1, a histone H2A variant that poises repressed/basal promoters for activation through histone loss. Cell 123, 219–231 (2005).
Struhl, K. Naturally occurring poly(dA-dT) sequences are upstream promoter elements for constitutive transcription in yeast. Proc. Natl Acad. Sci. USA 82, 8419–8423 (1985).
Lowary, P. T. & Widom, J. New DNA sequence rules for high affinity binding to histone octamer and sequence-directed nucleosome positioning. J. Mol. Biol. 276, 19–42 (1998).
Mavrich, T. N. et al. A barrier nucleosome model for statistical positioning of nucleosomes throughout the yeast genome. Genome Res. 18, 1073–1083 (2008).
Almer, A., Rudolph, H., Hinnen, A. & Horz, W. Removal of positioned nucleosomes from the yeast PHO5 promoter upon PHO5 induction releases additional upstream activating DNA elements. EMBO J. 5, 2689–2696 (1986).
Fascher, K. D., Schmitz, J. & Horz, W. Role of trans-activating proteins in the generation of active chromatin at the PHO5 promoter in S. cerevisiae . EMBO J. 9, 2523–2528 (1990).
Lam, F. H., Steger, D. J. & O'Shea, E. K. Chromatin decouples promoter threshold from dynamic range. Nature 453, 246–250 (2008).
Hebbar, P. B. & Archer, T. K. Chromatin remodeling by nuclear receptors. Chromosoma 111, 495–504 (2003).
Basehoar, A. D., Zanton, S. J. & Pugh, B. F. Identification and distinct regulation of yeast TATA box-containing genes. Cell 116, 699–709 (2004).
Huisinga, K. L. & Pugh, B. F. A genome-wide housekeeping role for TFIID and a highly regulated stress-related role for SAGA in Saccharomyces cerevisiae . Mol. Cell 13, 573–585 (2004).
Martinez-Campa, C. et al. Precise nucleosome positioning and the TATA box dictate requirements for the histone H4 tail and the bromodomain factor Bdf1. Mol. Cell 15, 69–81 (2004).
Grant, P. A., Sterner, D. E., Duggan, L. J., Workman, J. L. & Berger, S. L. The SAGA unfolds: convergence of transcription regulators in chromatin-modifying complexes. Trends Cell Biol. 8, 193–197 (1998).
Kingston, R. E. & Narlikar, G. J. ATP-dependent remodeling and acetylation as regulators of chromatin fluidity. Genes Dev. 13, 2339–2352 (1999).
Owen-Hughes, T. Colworth memorial lecture. Pathways for remodelling chromatin. Biochem Soc. Trans. 31, 893–905 (2003).
Langst, G., Bonte, E. J., Corona, D. F. & Becker, P. B. Nucleosome movement by CHRAC and ISWI without disruption or trans-displacement of the histone octamer. Cell 97, 843–852 (1999).
Ito, T., Bulger, M., Pazin, M. J., Kobayashi, R. & Kadonaga, J. T. ACF, an ISWI-containing and ATP-utilizing chromatin assembly and remodeling factor. Cell 90, 145–155 (1997).
Whitehouse, I. et al. Nucleosome mobilization catalysed by the yeast SWI/SNF complex. Nature 400, 784–787 (1999).
Lorch, Y., Zhang, M. & Kornberg, R. D. Histone octamer transfer by a chromatin-remodeling complex. Cell 96, 389–392 (1999). This paper shows that a chromatin remodeller can eject a nucleosome.
Mizuguchi, G. et al. ATP-driven exchange of histone H2AZ variant catalyzed by SWR1 chromatin remodeling complex. Science 303, 343–348 (2004). This paper provides the first evidence for in vitro insertion of H2A.Z by a remodeller.
Corona, D. F., Clapier, C. R., Becker, P. B. & Tamkun, J. W. Modulation of ISWI function by site-specific histone acetylation. EMBO Rep. 3, 242–247 (2002).
Kagalwala, M. N., Glaus, B. J., Dang, W., Zofall, M. & Bartholomew, B. Topography of the ISW2–nucleosome complex: insights into nucleosome spacing and chromatin remodeling. EMBO J. 23, 2092–2104 (2004).
Gelbart, M. E., Bachman, N., Delrow, J., Boeke, J. D. & Tsukiyama, T. Genome-wide identification of Isw2 chromatin-remodeling targets by localization of a catalytically inactive mutant. Genes Dev. 19, 942–954 (2005).
Whitehouse, I., Rando, O. J., Delrow, J. & Tsukiyama, T. Chromatin remodelling at promoters suppresses antisense transcription. Nature 450, 1031–1035 (2007).
Whitehouse, I. & Tsukiyama, T. Antagonistic forces that position nucleosomes in vivo . Nature Struct. Mol. Biol. 13, 633–640 (2006).
Parnell, T. J., Huff, J. T. & Cairns, B. R. RSC regulates nucleosome positioning at Pol II genes and density at Pol III genes. EMBO J. 27, 100–110 (2008).
Badis, G. et al. A library of yeast transcription factor motifs reveals a widespread function for Rsc3 in targeting nucleosome exclusion at promoters. Mol. Cell 32, 878–887 (2008).
Hartley, P. D. & Madhani, H. D. Mechanisms that specify promoter nucleosomes and identity. Cell 137, 445–458 (2009).
Henikoff, S. Nucleosome destabilization in the epigenetic regulation of gene expression. Nature Rev. Genet. 9, 15–26 (2008).
Keogh, M. C. et al. The Saccharomyces cerevisiae histone H2A variant Htz1 is acetylated by NuA4. Genes Dev. 20, 660–665 (2006).
Millar, C. B., Xu, F., Zhang, K. & Grunstein, M. Acetylation of H2AZ Lys 14 is associated with genome-wide gene activity in yeast. Genes Dev. 20, 711–722 (2006).
Jin, C. & Felsenfeld, G. Nucleosome stability mediated by histone variants H3.3 and H2A.Z. Genes Dev. 21, 1519–1529 (2007).
Ahmad, K. & Henikoff, S. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell 9, 1191–1200 (2002).
Raser, J. M. & O'Shea, E. K. Control of stochasticity in eukaryotic gene expression. Science 304, 1811–1814 (2004). This important paper connects chromatin attributes to transcriptional noise.
Raser, J. M. & O'Shea, E. K. Noise in gene expression: origins, consequences, and control. Science 309, 2010–2013 (2005).
Kaplan, C. D., Laprade, L. & Winston, F. Transcription elongation factors repress transcription initiation from cryptic sites. Science 301, 1096–1099 (2003).
Carrozza, M. J. et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell 123, 581–592 (2005).
Kornberg, R. D. & Stryer, L. Statistical distributions of nucleosomes: nonrandom locations by a stochastic mechanism. Nucleic Acids Res. 16, 6677–6690 (1988).
Core, L. J. & Lis, J. T. Transcription regulation through promoter-proximal pausing of RNA polymerase II. Science 319, 1791–1792 (2008).
Jiang, C. & Pugh, B. F. Nucleosome positioning and gene regulation: advances through genomics. Nature Rev. Genet. 10, 161–172 (2009).
Dion, M. F. et al. Dynamics of replication-independent histone turnover in budding yeast. Science 315, 1405–1408 (2007).
Rufiange, A., Jacques, P. E., Bhat, W., Robert, F. & Nourani, A. Genome-wide replication-independent histone H3 exchange occurs predominantly at promoters and implicates H3 K56 acetylation and Asf1. Mol. Cell 27, 393–405 (2007).
Adkins, M. W., Howar, S. R. & Tyler, J. K. Chromatin disassembly mediated by the histone chaperone Asf1 is essential for transcriptional activation of the yeast PHO5 and PHO8 genes. Mol. Cell 14, 657–666 (2004).
Acknowledgements
I thank T. Parnell and C. Clapier for comments and assistance with figures. I am grateful for support from the US National Institutes of Health (grant GM60415) and the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Ethics declarations
Competing interests
The author declares no competing financial interests.
Additional information
Reprints and permissions information is available at http://www.nature.com/reprints. The author declares no competing financial interests. Correspondence should be addressed to B.R.C. (brad.cairns@hci.utah.edu).
Rights and permissions
About this article
Cite this article
Cairns, B. The logic of chromatin architecture and remodelling at promoters. Nature 461, 193–198 (2009). https://doi.org/10.1038/nature08450
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature08450
This article is cited by
-
Regulation of the RNA polymerase II pre-initiation complex by its associated coactivators
Nature Reviews Genetics (2023)
-
Trps1 acts as a regulator of Sf-1 transcription and testosterone synthesis in mouse Leydig cells
Cell Biology and Toxicology (2023)
-
SUPPRESSOR OF PHYTOCHROME B-4 #3 reduces the expression of PIF-activated genes and increases expression of growth repressors to regulate hypocotyl elongation in short days
BMC Plant Biology (2022)
-
Emerging roles of SWI/SNF remodelers in fungal pathogens
Current Genetics (2022)
-
Production of a viral surface protein in Nannochloropsis oceanica for fish vaccination against infectious pancreatic necrosis virus
Applied Microbiology and Biotechnology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.