Abstract
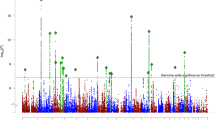
Schizophrenia is a severe mental disorder with a lifetime risk of about 1%, characterized by hallucinations, delusions and cognitive deficits, with heritability estimated at up to 80%1,2. We performed a genome-wide association study of 3,322 European individuals with schizophrenia and 3,587 controls. Here we show, using two analytic approaches, the extent to which common genetic variation underlies the risk of schizophrenia. First, we implicate the major histocompatibility complex. Second, we provide molecular genetic evidence for a substantial polygenic component to the risk of schizophrenia involving thousands of common alleles of very small effect. We show that this component also contributes to the risk of bipolar disorder, but not to several non-psychiatric diseases.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
06 August 2009
The subheading for Table 1b was changed on 6 August 2009.
References
Cardno, A. G. & Gottesman, I. I. Twin studies of schizophrenia: from bow-and-arrow concordances to star wars Mx and functional genomics. Am. J. Med. Genet. 97, 12–17 (2000)
Sullivan, P. F., Kendler, K. S. & Neale, M. C. Schizophrenia as a complex trait: evidence from a meta-analysis of twin studies. Arch. Gen. Psychiatry 60, 1187–1192 (2003)
O’Donovan, M. C. et al. Identification of loci associated with schizophrenia by genome-wide association and follow-up. Nature Genet. 40, 1053–1055 (2008)
Wei, J. & Hemmings, G. P. The NOTCH4 locus is associated with susceptibility to schizophrenia. Nature Genet. 25, 376–377 (2000)
Horton, R. et al. Variation analysis and gene annotation of eight MHC haplotypes: the MHC Haplotype Project. Immunogenetics 60, 1–18 (2008)
Stefansson, H. et al. Common variants conferring risk of schizophrenia. Nature 10.1038/nature08186 (this issue)
Douglas, J. S. et al. Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature 10.1038/nature08192 (this issue)
Fisher, R. A. The correlation between relatives on the supposition of Mendelian inheritance. Philos. Trans. R. Soc. Edinb. 52, 399–433 (1918)
Gottesman, I. I. & Shields, J. A polygenic theory of schizophrenia. Proc. Natl Acad. Sci. USA 58, 199–205 (1967)
Wray, N. R., Goddard, M. E. & Visscher, P. M. Prediction of individual genetic risk to disease from genome-wide association studies. Genome Res. 17, 1520–1528 (2007)
Craddock, N., O’Donovan, M. C. & Owen, M. J. Genes for schizophrenia and bipolar disorder? Implications for psychiatric nosology. Schizophr. Bull. 32, 9–16 (2006)
Sklar, P. et al. Whole-genome association study of bipolar disorder. Mol. Psychiatry 13, 558–569 (2008)
The Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447, 661–678 (2007)
Svensson, A. C., Lichtenstein, P., Sandin, S. & Hultman, C. M. Fertility of first-degree relatives of patients with schizophrenia: a three generation perspective. Schizophr. Res. 91, 238–245 (2007)
McClellan, J. M., Susser, E. & King, M. C. Schizophrenia: a common disease caused by multiple rare alleles. Br. J. Psychiatry 190, 194–199 (2007)
Craddock, N., O’Donovan, M. C. & Owen, M. J. Phenotypic and genetic complexity of psychosis. Invited commentary on. Schizophrenia: a common disease caused by multiple rare alleles. Br. J. Psychiatry 190, 200–203 (2007)
International Schizophrenia Consortium. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature 455, 237–241 (2008)
Walsh, T. et al. Rare structural variants disrupt multiple genes in neurodevelopmental pathways in schizophrenia. Science 320, 539–543 (2008)
Xu, B. et al. Strong association of de novo copy number mutations with sporadic schizophrenia. Nature Genet. 40, 880–885 (2008)
Weedon, M. N. et al. Genome-wide association analysis identifies 20 loci that influence adult height. Nature Genet. 40, 575–583 (2008)
Lichtenstein, P. et al. Common genetic determinants of schizophrenia and bipolar disorder in Swedish families: a population-based study. Lancet 373, 234–239 (2009)
Psychiatric GWAS Consortium Steering Committee. A framework for interpreting genome-wide association studies of psychiatric disorders. Mol. Psychiatry 14, 10–17 (2009)
Psychiatric GWAS Consortium Coordinating Committee. Genomewide association studies: history, rationale and prospects for psychiatric disorders. Am. J. Psychiatry 166, 540–556 (2009)
Cross Disorder Phenotype Group of the Psychiatric GWAS Consortium Dissecting the phenotype in genome-wide association studies of psychiatric illness. Br. J. Psychiatry 10.1192/bjp.bp.108.063156 (in the press)
Manolio, T. A., Brooks, L. D. & Collins, F. S. A. HapMap harvest of insights into the genetics of common disease. J. Clin. Invest. 118, 1590–1605 (2008)
Maher, B. The case of the missing heritability. Nature 456, 18–21 (2008)
Korn, J. M. et al. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nature Genet. 40, 1253–1260 (2008)
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007)
Acknowledgements
We thank the patients and families who contributed to these studies. We also thank E. Lander, N. Patterson and members of the Medical and Population Genetics group at the Broad Institute of Harvard and Massachusetts Institute of Technology for valuable discussion, and members of the Broad Biological Samples and Genetic Analysis Platforms for sample management and genotyping. We particularly thank D. Levinson and P. Gejman for allowing access to the MGS samples, and J. Shi for analytic support with the MGS samples. The group at the Stanley Center for Psychiatric Research at the Broad Institute was supported by the Stanley Medical Research Institute (E.M.S.), the Sylvan C. Herman Foundation (E.M.S.), and MH071681 (P.S.). The Cardiff University group was supported by a Medical Research Council (UK) Programme grant and the National Institutes of Mental Health (USA) (CONTE: 2 P50 MH066392-05A1). The group at the Karolinska Institutet was supported by the Swedish Council for Working Life and Social Research (FO 184/2000; 2001-2368). The Massachusetts General Hospital group was supported by the Stanley Medical Research Institute (P.S.), MH071681 and MH077139 (P.S.) and a Narsad Young Investigator Award (S.M.P.). The group at the Queensland Institute of Medical Research was supported by the Australian National Health and Medical Research Council (grants 389892, 442915, 496688 and 496674) and thanks S. Gordon for data preparation. The Trinity College Dublin group was supported by Science Foundation Ireland, the Health Research Board (Ireland), the Stanley Medical Research Institute and the Wellcome Trust; Irish controls were supplied by J. McPartlin from the Trinity College Biobank. The work at the University of Aberdeen was partly funded by GlaxoSmithKline and Generation Scotland, Genetics Health Initiative. University College London clinical and control samples were collected with support from the Neuroscience Research Charitable Trust, the Camden and Islington Mental Health and Social Care Trust, East London and City Mental Heath Trust, the West Berkshire NHS Trust, the West London Mental Health Trust, Oxfordshire and Buckinghamshire Mental Health Partnership NHS Trust, South Essex Partnership NHS Foundation Trust, Gloucestershire Partnership NHS Foundation Trust, Mersey Care NHS Trust, Hampshire Partnership NHS Trust and the North East London Mental Health Trust. The collection of the University of Edinburgh cohort was supported by the Wellcome Trust Clinical Research Facility (Edinburgh) and grants from The Wellcome Trust, London and the Chief Scientist Office of the Scottish Government. The group at the University of North Carolina, Chapel Hill, was supported by MH074027, MH077139 and MH080403, the Sylvan C. Herman Foundation (P.F.S.) and the Stanley Medical Research Institute (P.F.S.). The group at the University of Southern California thanks the patients and their families for their collaboration, and acknowledges the support of the National Institutes of Mental Health and the Department of Veterans Affairs.
Author information
Authors and Affiliations
Consortia
Corresponding authors
Supplementary information
Supplementary Information
This file contains Supplementary Notes and Data with Supplementary Tables 1-18, Supplementary Figures 1-8 with Legends and Supplementary References. (PDF 2178 kb)
Rights and permissions
About this article
Cite this article
The International Schizophrenia Consortium. Common polygenic variation contributes to risk of schizophrenia and bipolar disorder. Nature 460, 748–752 (2009). https://doi.org/10.1038/nature08185
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature08185
This article is cited by
-
SNVstory: inferring genetic ancestry from genome sequencing data
BMC Bioinformatics (2024)
-
Patients with first-episode psychosis in northern Taiwan: neurocognitive performance and niacin response profile in comparison with schizophrenia patients of different familial loadings and relationship with clinical features
BMC Psychiatry (2024)
-
Genetic influences on circulating retinol and its relationship to human health
Nature Communications (2024)
-
Principles and methods for transferring polygenic risk scores across global populations
Nature Reviews Genetics (2024)
-
Exploring functional dysconnectivity in schizophrenia: alterations in eigenvector centrality mapping and insights into related genes from transcriptional profiles
Schizophrenia (2024)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.