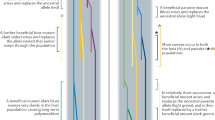
Abstract
The ecology, behaviour and genetics of our closest living relatives, the nonhuman primates, should help us to understand the evolution of our own lineage. Although a large amount of data has been amassed on primate ecology and behaviour, much less is known about the functional and evolutionary genetic aspects of primate biology, especially in wild primates. As a result, even in well-studied populations in which nongenetic factors that influence adaptively important characteristics have been identified, we have almost no understanding of the underlying genetic basis for such traits. Here, we report on the functional consequences of genetic variation at the malaria-related FY (DARC) gene in a well-studied population of yellow baboons (Papio cynocephalus) living in Amboseli National Park in Kenya. FY codes for a chemokine receptor normally expressed on the erythrocyte surface that is the known entry point for the malarial parasite Plasmodium vivax1,2,3. We identified variation in the cis-regulatory region of the baboon FY gene that was associated with phenotypic variation in susceptibility to Hepatocystis, a malaria-like pathogen that is common in baboons4,5. Genetic variation in this region also influenced gene expression in vivo in wild individuals, a result we confirmed using in vitro reporter gene assays. The patterns of genetic variation in and around this locus were also suggestive of non-neutral evolution, raising the possibility that the evolution of the FY cis-regulatory region in baboons has exhibited both mechanistic and selective parallels with the homologous region in humans6,7,8. Together, our results represent the first reported association and functional characterization linking genetic variation and a complex trait in a natural population of nonhuman primates.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Data deposits
Sequence data have been deposited in NCBI GenBank under the accession numbers FJ952954–FJ955880, FJ955882–FJ955885, FJ955887–FJ955896 and FJ955899–FJ956699.
References
Barnwell, J. W., Nichols, M. E. & Rubinstein, P. In vitro evaluation of the role of the Duffy blood group in erythrocyte invasion by Plasmodium vivax. J. Exp. Med. 169, 1795–1802 (1989)
Miller, L. H., Mason, S. J., Clyde, D. F. & Mcginniss, M. H. Resistance factor to Plasmodium vivax in blacks—Duffy blood group genotype Fyfy. New Engl. J. Med. 295, 302–304 (1976)
Miller, L. H., Mason, S. J., Dvorak, J. A., Mcginniss, M. H. & Rothman, I. K. Erythrocyte receptors for Plasmodium knowlesi malaria—Duffy blood group determinants. Science 189, 561–563 (1975)
Garnham, P. C. C. Malaria Parasites and Other Haemosporidia (Blackwell, 1966)
Myers, B. J. & Kuntz, R. E. A checklist of parasites reported for the baboon. Primates 6, 137–194 (1965)
Hamblin, M. T. & Di Rienzo, A. Detection of the signature of natural selection in humans: evidence from the Duffy blood group locus. Am. J. Hum. Genet. 66, 1669–1679 (2000)
Hamblin, M. T., Thompson, E. E. & Di Rienzo, A. Complex signatures of natural selection at the Duffy blood group locus. Am. J. Hum. Genet. 70, 369–383 (2002)
Sabeti, P. C. et al. Positive natural selection in the human lineage. Science 312, 1614–1620 (2006)
Tournamille, C., Colin, Y., Cartron, J. P. & Levankim, C. Disruption of a GATA motif in the Duffy gene promoter abolishes erythroid gene expression in Duffy negative individuals. Nature Genet. 10, 224–228 (1995)
Michon, P. et al. Duffy-null promoter heterozygosity reduces DARC expression and abrogates adhesion of the P. vivax ligand required for blood-stage infection. FEBS Lett. 495, 111–114 (2001)
Zimmerman, P. A. et al. Emergence of FY*A(null) in a Plasmodium vivax-endemic region of Papua New Guinea. Proc. Natl Acad. Sci. USA 96, 13973–13977 (1999)
Alberts, S. C., Buchan, J. C. & Altmann, J. Sexual selection in wild baboons: from mating opportunities to paternity success. Anim. Behav. 72, 1177–1196 (2006)
Buchan, J. C., Alberts, S. C., Silk, J. B. & Altmann, J. True paternal care in a multi-male primate society. Nature 425, 179–181 (2003)
Tung, J., Charpentier, M. J. E., Garfield, D. A., Altmann, J. & Alberts, S. C. Genetic evidence reveals temporal change in hybridization patterns in a wild baboon population. Mol. Ecol. 17, 1998–2011 (2008)
Perkins, S. L. & Schall, J. J. A molecular phylogeny of malarial parasites recovered from cytochrome b gene sequences. J. Parasitol. 88, 972–978 (2002)
Garnham, P. C. C., Heisch, R. B., Minter, D. M., Phipps, J. D. & Ikata, M. Culicoides adersi Ingram and Macfie, 1923, a presumed vector of Hepatocystis ( = Plasmodium) kochi (Laveran, 1899). Nature 190, 739–741 (1961)
Wittkopp, P., Haerum, B. & Clark, A. Evolutionary changes in cis and trans gene regulation. Nature 430, 85–88 (2004)
Yan, H., Yuan, W., Velculescu, V. E., Vogelstein, B. & Kinzler, K. W. Allelic variation in human gene expression. Science 297, 1143 (2002)
Reich, D. et al. Reduced neutrophil count in people of African descent is due to a regulatory variant in the Duffy antigen receptor for chemokines gene. PLoS Genet. 5, e1000360 (2009)
Loisel, D. A., Rockman, M. V., Wray, G. A., Altmann, J. & Alberts, S. C. Ancient polymorphism and functional variation in the primate MHC-DQA1 5' cis-regulatory region. Proc. Natl Acad. Sci. USA 103, 16331–16336 (2006)
Wooding, S. et al. Independent evolution of bitter-taste sensitivity in humans and chimpanzees. Nature 440, 930–934 (2006)
Loisel, D. Evolutionary Genetics of Immune System Genes in a Wild Primate Population. PhD thesis, Duke Univ. (2007)
Seixas, S., Ferrand, N. & Rocha, J. Microsatellite variation and evolution of the human Duffy blood group polymorphism. Mol. Biol. Evol. 19, 1802–1806 (2002)
Moore, J. A. & Kuntz, R. E. Entopolyploides macaci Mayer, 1934 in the African baboon (Papio cynocephalus L. 1766). J. Med. Primatol. 4, 1–7 (1975)
Brem, R. B., Storey, J. D., Whittle, J. & Kruglyak, L. Genetic interactions between polymorphisms that affect gene expression in yeast. Nature 436, 701–703 (2005)
Smith, E. N. & Kruglyak, L. Gene-environment interaction in yeast gene expression. PLoS Biol. 6, e83 (2008)
Rozas, J., Sanchez-Del Barrio, J. C., Messeguer, X., Rozas, R. & Dna, S. P. DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19, 2496–2497 (2003)
Altmann, J. et al. Behavior predicts genetic structure in a wild primate group. Proc. Natl Acad. Sci. USA 93, 5797–5801 (1996)
Stephens, M., Smith, N. J. & Donnelly, P. A new statistical method for haplotype reconstruction from population data. Am. J. Hum. Genet. 68, 978–989 (2001)
Excoffier, L., Laval, G. & Schneider, S. Arlequin version 3.0: an integrated software package for population genetics data analysis. Evol. Bioinform. Online 1, 47–50 (2005)
Acknowledgements
We thank the Office of the President of the Republic of Kenya and the Kenya Wildlife Service for permission to work in Amboseli National Park, and the Institute of Primate Research for local sponsorship. We thank the wardens and staff of Amboseli National Park, and the pastoral communities of Amboseli and Longido for cooperation. We thank J. Altmann for providing access to long-term data and contributing samples, and J. Altmann and Y. Gilad for providing comments on the manuscript. R. S. Mututua, S. Sayialel, and J. K. Warutere assisted with sample collection. The Integrated Primate Biomaterials and Information Resource, the Coriell Institute, J. Rogers and R. Sapolsky provided access to DNA samples from Mikumi and Masai Mara respectively. G. Gibson, T. F. C. Mackay, L. Goering and D. Tan provided access to a pyrosequencer at NC State University. M. Akinyi assisted with sample collection and analysis. A. D. Pfefferle assisted with sequencing. S. Mukherjee advised and assisted with statistical tests. Financial support came from the National Science Foundation (to S.C.A. and J.T.); the American Society of Primatologists (to J.T.); Duke University and the Duke chapter of Sigma Xi (to J.T.); and the Duke Institute for Genome Sciences and Policy (to G.A.W.).
Author Contributions J.T., S.C.A. and G.A.W. designed the study, analysed the results, and wrote the paper. J.T. and S.C.A. collected blood samples; S.C.A. provided the long-term data on Amboseli National Park. J.T. gathered the allelic imbalance and sequence data; A.P., T.F.S. and J.T. collected the transfection assay data; T.F.S. and J.T. collected the genotyping data; A.J.B. and J.T. collected the Hepatocystis data. G.A.W. and S.C.A. provided funding support.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-4 with Legends, Supplementary Methods, Supplementary Tables 1-4 and Supplementary References. (PDF 641 kb)
Rights and permissions
About this article
Cite this article
Tung, J., Primus, A., Bouley, A. et al. Evolution of a malaria resistance gene in wild primates. Nature 460, 388–391 (2009). https://doi.org/10.1038/nature08149
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature08149
This article is cited by
-
Epauletted fruit bats display exceptionally high infections with a Hepatocystis species complex in South Sudan
Scientific Reports (2017)
-
Primate genotyping via high resolution melt analysis: rapid and reliable identification of color vision status in wild lemurs
Primates (2016)
-
Immune regulation by atypical chemokine receptors
Nature Reviews Immunology (2013)
-
Resistance to malaria in humans: the impact of strong, recent selection
Malaria Journal (2012)
-
Molecular evolution of a malaria resistance gene (DARC) in primates
Immunogenetics (2012)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.