Abstract
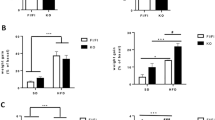
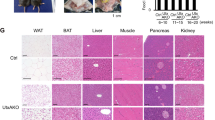
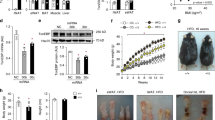
Recent studies indicate that the methylation state of histones can be dynamically regulated by histone methyltransferases and demethylases1,2. The H3K9-specific demethylase Jhdm2a (also known as Jmjd1a and Kdm3a) has an important role in nuclear hormone receptor-mediated gene activation and male germ cell development3,4. Through disruption of the Jhdm2a gene in mice, here we demonstrate that Jhdm2a is critically important in regulating the expression of metabolic genes. The loss of Jhdm2a function results in obesity and hyperlipidemia in mice. We provide evidence that the loss of Jhdm2a function disrupts β-adrenergic-stimulated glycerol release and oxygen consumption in brown fat, and decreases fat oxidation and glycerol release in skeletal muscles. We show that Jhdm2a expression is induced by β-adrenergic stimulation, and that Jhdm2a directly regulates peroxisome proliferator-activated receptor α (Ppara) and Ucp1 expression. Furthermore, we demonstrate that β-adrenergic activation-induced binding of Jhdm2a to the PPAR responsive element (PPRE) of the Ucp1 gene not only decreases levels of H3K9me2 (dimethylation of lysine 9 of histone H3) at the PPRE, but also facilitates the recruitment of Pparγ and Rxrα and their co-activators Pgc1α (also known as Ppargc1a), CBP/p300 (Crebbp) and Src1 (Ncoa1) to the PPRE. Our studies thus demonstrate an essential role for Jhdm2a in regulating metabolic gene expression and normal weight control in mice.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Klose, R. J. & Zhang, Y. Regulation of histone methylation by demethylimination and demethylation. Nature Rev. Mol. Cell Biol. 8, 307–318 (2007)
Martin, C. & Zhang, Y. The diverse functions of histone lysine methylation. Nature Rev. Mol. Cell Biol. 6, 838–849 (2005)
Okada, Y., Scott, G., Ray, M. K., Mishina, Y. & Zhang, Y. Histone demethylase JHDM2A is critical for Tnp1 and Prm1 transcription and spermatogenesis. Nature 450, 119–123 (2007)
Yamane, K. et al. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell 125, 483–495 (2006)
Spiegelman, B. M. & Flier, J. S. Obesity and the regulation of energy balance. Cell 104, 531–543 (2001)
Evans, R. M., Barish, G. D. & Wang, Y. X. PPARs and the complex journey to obesity. Nature Med. 10, 355–361 (2004)
Reddy, J. K. & Hashimoto, T. Peroxisomal β-oxidation and peroxisome proliferator-activated receptor alpha: an adaptive metabolic system. Annu. Rev. Nutr. 21, 193–230 (2001)
Bedu, E., Desplanches, D., Pequignot, J., Bordier, B. & Desvergne, B. Double gene deletion reveals the lack of cooperation between PPARα and PPARβ in skeletal muscle. Biochem. Biophys. Res. Commun. 357, 877–881 (2007)
Finck, B. N. et al. A potential link between muscle peroxisome proliferator- activated receptor-α signaling and obesity-related diabetes. Cell Metab. 1, 133–144 (2005)
Hibuse, T. et al. Aquaporin 7 deficiency is associated with development of obesity through activation of adipose glycerol kinase. Proc. Natl Acad. Sci. USA 102, 10993–10998 (2005)
Gulick, T., Cresci, S., Caira, T., Moore, D. D. & Kelly, D. P. The peroxisome proliferator-activated receptor regulates mitochondrial fatty acid oxidative enzyme gene expression. Proc. Natl Acad. Sci. USA 91, 11012–11016 (1994)
Lopez, D., Irby, R. B. & McLean, M. P. Peroxisome proliferator-activated receptor α induces rat sterol carrier protein x promoter activity through two peroxisome proliferator-response elements. Mol. Cell. Endocrinol. 205, 169–184 (2003)
Pineda Torra, I., Jamshidi, Y., Flavell, D. M., Fruchart, J. C. & Staels, B. Characterization of the human PPARα promoter: identification of a functional nuclear receptor response element. Mol. Endocrinol. 16, 1013–1028 (2002)
Tugwood, J. D. et al. The mouse peroxisome proliferator activated receptor recognizes a response element in the 5′ flanking sequence of the rat acyl CoA oxidase gene. EMBO J. 11, 433–439 (1992)
Bachman, E. S. et al. βAR signaling required for diet-induced thermogenesis and obesity resistance. Science 297, 843–845 (2002)
Lowell, B. B. & Spiegelman, B. M. Towards a molecular understanding of adaptive thermogenesis. Nature 404, 652–660 (2000)
Enerback, S. et al. Mice lacking mitochondrial uncoupling protein are cold-sensitive but not obese. Nature 387, 90–94 (1997)
Kersten, S. et al. Peroxisome proliferator-activated receptor α mediates the adaptive response to fasting. J. Clin. Invest. 103, 1489–1498 (1999)
Cassard-Doulcier, A. M. et al. Tissue-specific and β-adrenergic regulation of the mitochondrial uncoupling protein gene: control by cis-acting elements in the 5′-flanking region. Mol. Endocrinol. 7, 497–506 (1993)
Wang, Z. et al. Critical roles of the p160 transcriptional coactivators p/CIP and SRC-1 in energy balance. Cell Metab. 3, 111–122 (2006)
Lomax, M. A. et al. Ontogenic loss of brown adipose tissue sensitivity to beta-adrenergic stimulation in the ovine. Endocrinology 148, 461–468 (2007)
Rando, T. A. & Blau, H. M. Primary mouse myoblast purification, characterization, and transplantation for cell-mediated gene therapy. J. Cell Biol. 125, 1275–1287 (1994)
Ross, S. R. et al. Hibernoma formation in transgenic mice and isolation of a brown adipocyte cell line expressing the uncoupling protein gene. Proc. Natl Acad. Sci. USA 89, 7561–7565 (1992)
Acknowledgements
We thank B. M. Spiegelman for the HIB1B cells, L. Xia for construction of the targeting vector, K. E. Gardner for critical reading of the manuscript, D. Pump and K. Hua (UNC Clinical Nutrition Research Unit, DK56350) for calorimetry and MRI, and N. Takahashi for helpful comments. Y.Z. is an investigator of the Howard Hughes Medical Institute.
Author Contributions K.T. and Y.Z. designed the experiments and prepared the manuscript. K.T. performed most of the experiments. Y.O. provided the data for Supplementary Fig. 3. E.K. analysed microarray data and generated Supplementary Figs 6 and 7.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Information
This file contains Supplementary Methods, Supplementary References, Supplementary Figures S1-S12 and Supplementary Table 1. (PDF 5889 kb)
Rights and permissions
About this article
Cite this article
Tateishi, K., Okada, Y., Kallin, E. et al. Role of Jhdm2a in regulating metabolic gene expression and obesity resistance. Nature 458, 757–761 (2009). https://doi.org/10.1038/nature07777
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature07777
This article is cited by
-
Mesenchymal stem cells under epigenetic control – the role of epigenetic machinery in fate decision and functional properties
Cell Death & Disease (2023)
-
Epitranscriptomics in metabolic disease
Nature Metabolism (2023)
-
Embryonic vitamin D deficiency programs hematopoietic stem cells to induce type 2 diabetes
Nature Communications (2023)
-
Epigenetics and the role of nutraceuticals in health and disease
Environmental Science and Pollution Research (2023)
-
HIF-1α/JMJD1A signaling regulates inflammation and oxidative stress following hyperglycemia and hypoxia-induced vascular cell injury
Cellular & Molecular Biology Letters (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.