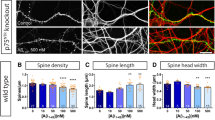
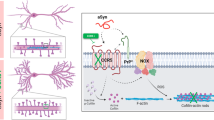
Abstract
A pathological hallmark of Alzheimer’s disease is an accumulation of insoluble plaque containing the amyloid-β peptide of 40–42 amino acid residues1. Prefibrillar, soluble oligomers of amyloid-β have been recognized to be early and key intermediates in Alzheimer’s-disease-related synaptic dysfunction2,3,4,5,6,7,8,9. At nanomolar concentrations, soluble amyloid-β oligomers block hippocampal long-term potentiation7, cause dendritic spine retraction from pyramidal cells5,8 and impair rodent spatial memory2. Soluble amyloid-β oligomers have been prepared from chemical syntheses, transfected cell culture supernatants, transgenic mouse brain and human Alzheimer’s disease brain2,4,7,9. Together, these data imply a high-affinity cell-surface receptor for soluble amyloid-β oligomers on neurons—one that is central to the pathophysiological process in Alzheimer’s disease. Here we identify the cellular prion protein (PrPC) as an amyloid-β-oligomer receptor by expression cloning. Amyloid-β oligomers bind with nanomolar affinity to PrPC, but the interaction does not require the infectious PrPSc conformation. Synaptic responsiveness in hippocampal slices from young adult PrP null mice is normal, but the amyloid-β oligomer blockade of long-term potentiation is absent. Anti-PrP antibodies prevent amyloid-β-oligomer binding to PrPC and rescue synaptic plasticity in hippocampal slices from oligomeric amyloid-β. Thus, PrPC is a mediator of amyloid-β-oligomer-induced synaptic dysfunction, and PrPC-specific pharmaceuticals may have therapeutic potential for Alzheimer’s disease.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Hardy, J. & Selkoe, D. J. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science 297, 353–356 (2002)
Lesne, S. et al. A specific amyloid-β protein assembly in the brain impairs memory. Nature 440, 352–357 (2006)
Cleary, J. P. et al. Natural oligomers of the amyloid-β protein specifically disrupt cognitive function. Nature Neurosci. 8, 79–84 (2005)
Chromy, B. A. et al. Self-assembly of Aβ1–42 into globular neurotoxins. Biochemistry 42, 12749–12760 (2003)
Lacor, P. N. et al. Abeta oligomer-induced aberrations in synapse composition, shape, and density provide a molecular basis for loss of connectivity in Alzheimer’s disease. J. Neurosci. 27, 796–807 (2007)
Lacor, P. N. et al. Synaptic targeting by Alzheimer’s-related amyloid β oligomers. J. Neurosci. 24, 10191–10200 (2004)
Walsh, D. M. et al. Naturally secreted oligomers of amyloid β protein potently inhibit hippocampal long-term potentiation in vivo . Nature 416, 535–539 (2002)
Shankar, G. M. et al. Natural oligomers of the Alzheimer amyloid-β protein induce reversible synapse loss by modulating an NMDA-type glutamate receptor-dependent signaling pathway. J. Neurosci. 27, 2866–2875 (2007)
Shankar, G. M. et al. Amyloid-β protein dimers isolated directly from Alzheimer’s brains impair synaptic plasticity and memory. Nature Med. 14, 837–842 (2008)
Hepler, R. W. et al. Solution state characterization of amyloid β-derived diffusible ligands. Biochemistry 45, 15157–15167 (2006)
Lambert, M. P. et al. Diffusible, nonfibrillar ligands derived from Aβ1–42 are potent central nervous system neurotoxins. Proc. Natl Acad. Sci. USA 95, 6448–6453 (1998)
Prusiner, S. B. Prions. Proc. Natl Acad. Sci. USA 95, 13363–13383 (1998)
Yan, S. D. et al. RAGE and amyloid-β peptide neurotoxicity in Alzheimer’s disease. Nature 382, 685–691 (1996)
Wang, H. Y. et al. β-Amyloid1–42 binds to α7 nicotinic acetylcholine receptor with high affinity. Implications for Alzheimer’s disease pathology. J. Biol. Chem. 275, 5626–5632 (2000)
Viles, J. H. et al. Copper binding to the prion protein: structural implications of four identical cooperative binding sites. Proc. Natl Acad. Sci. USA 96, 2042–2047 (1999)
Jackson, G. S. et al. Location and properties of metal-binding sites on the human prion protein. Proc. Natl Acad. Sci. USA 98, 8531–8535 (2001)
Baumann, F. et al. Lethal recessive myelin toxicity of prion protein lacking its central domain. EMBO J. 26, 538–547 (2007)
Li, A. et al. Neonatal lethality in transgenic mice expressing prion protein with a deletion of residues 105–125. EMBO J. 26, 548–558 (2007)
Riek, R. et al. NMR structure of the mouse prion protein domain PrP(121–321). Nature 382, 180–182 (1996)
Bueler, H. et al. Normal development and behaviour of mice lacking the neuronal cell-surface PrP protein. Nature 356, 577–582 (1992)
Manson, J. C. et al. 129/Ola mice carrying a null mutation in PrP that abolishes mRNA production are developmentally normal. Mol. Neurobiol. 8, 121–127 (1994)
Lledo, P. M., Tremblay, P., DeArmond, S. J., Prusiner, S. B. & Nicoll, R. A. Mice deficient for prion protein exhibit normal neuronal excitability and synaptic transmission in the hippocampus. Proc. Natl Acad. Sci. USA 93, 2403–2407 (1996)
Curtis, J., Errington, M., Bliss, T., Voss, K. & MacLeod, N. Age-dependent loss of PTP and LTP in the hippocampus of PrP-null mice. Neurobiol. Dis. 13, 55–62 (2003)
Riemenschneider, M. et al. Prion protein codon 129 polymorphism and risk of Alzheimer disease. Neurology 63, 364–366 (2004)
Papassotiropoulos, A. et al. The prion gene is associated with human long-term memory. Hum. Mol. Genet. 14, 2241–2246 (2005)
Yehiely, F. et al. Identification of candidate proteins binding to prion protein. Neurobiol. Dis. 3, 339–355 (1997)
Schmitt-Ulms, G. et al. Time-controlled transcardiac perfusion cross-linking for the study of protein interactions in complex tissues. Nature Biotechnol. 22, 724–731 (2004)
Venkitaramani, D. V. et al. β-amyloid modulation of synaptic transmission and plasticity. J. Neurosci. 27, 11832–11837 (2007)
Hsieh, H. et al. AMPAR removal underlies Aβ-induced synaptic depression and dendritic spine loss. Neuron 52, 831–843 (2006)
Khosravani, H. et al. Prion protein attenuates excitotoxicity by inhibiting NMDA receptors. J. Cell Biol. 181, 551–565 (2008)
Folta-Stogniew, E. Oligomeric states of proteins determined by size-exclusion chromatography coupled with light scattering, absorbance, and refractive index detectors. Methods Mol. Biol. 328, 97–112 (2006)
Rajagopalan, S. et al. Neogenin mediates the action of repulsive guidance molecule. Nature Cell Biol. 6, 756–762 (2004)
Fournier, A. E., GrandPre, T. & Strittmatter, S. M. Identification of a receptor mediating Nogo-66 inhibition of axonal regeneration. Nature 409, 341–346 (2001)
Takahashi, T. et al. Plexin–neuropilin-1 complexes form functional semaphorin-3A receptors. Cell 99, 59–69 (1999)
Takahashi, T., Nakamura, F., Jin, Z., Kalb, R. G. & Strittmatter, S. M. Semaphorins A and E act as antagonists of neuropilin-1 and agonists of neuropilin-2 receptors. Nature Neurosci. 1, 487–493 (1998)
Weissmann, C. & Flechsig, E. PrP knock-out and PrP transgenic mice in prion research. Br. Med. Bull. 66, 43–60 (2003)
Chen, S., Mange, A., Dong, L., Lehmann, S. & Schachner, M. Prion protein as trans-interacting partner for neurons is involved in neurite outgrowth and neuronal survival. Mol. Cell. Neurosci. 22, 227–233 (2003)
Acknowledgements
We thank S. Tomita for cRNAs encoding GluR1–4 and stargazin (also known as CACNG2), B. Chesebro for providing us the Prnp null mice, M. Schachner for providing the PrP–Fc expression vector, E. Flechsig, C. Weismann and D. Harris for providing the PrPC deletion expression plasmids, D. Westaway for the Sprn expression plasmid and P. Seeburg for N-methyl-d-aspartate receptor subunit cDNAs. We thank S. Sodi for assistance with mouse husbandry. We thank E. Folta-Stogniew for SEC, and C. Rahner and M. Graham for electron microscopy. J.L. is a Brown-Coxe Postdoctoral Fellow, J.W.G. is supported by NIH Medical Scientist training Program grant 5T32GN07205, and S.M.S. is a member of the Kavli Institute for Neuroscience at Yale University. This work was supported by research grants from the Falk Medical Research Trust and the NIH to S.M.S. The SEC was supported by a NIDA-funded Neuroproteomic Center.
Author Contributions J.L. performed the amyloid-β binding and expression cloning experiments, D.A.G. conducted mouse breeding and tissue biochemistry, S.M.S. and H.B.N. performed the hippocampal electrophysiology experiments, and S.M.S., J.W.G. and J.L. performed the X. laevis studies. S.M.S. supervised all experiments. All authors participated in writing the manuscript.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Figures
This file contains Supplementary Figures 1-15 with Legends (PDF 3399 kb)
Rights and permissions
About this article
Cite this article
Laurén, J., Gimbel, D., Nygaard, H. et al. Cellular prion protein mediates impairment of synaptic plasticity by amyloid-β oligomers. Nature 457, 1128–1132 (2009). https://doi.org/10.1038/nature07761
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature07761
This article is cited by
-
Molecular Mechanisms Associated with Neurodegeneration of Neurotropic Viral Infection
Molecular Neurobiology (2024)
-
Synaptic degeneration in Alzheimer disease
Nature Reviews Neurology (2023)
-
Prion assemblies: structural heterogeneity, mechanisms of formation, and role in species barrier
Cell and Tissue Research (2023)
-
Soluble and insoluble protein aggregates, endoplasmic reticulum stress, and vascular dysfunction in Alzheimer’s disease and cardiovascular diseases
GeroScience (2023)
-
Non-human primates in prion diseases
Cell and Tissue Research (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.