Abstract
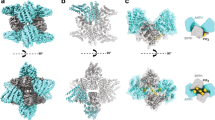
Most transmembrane proteins are selected as transport-vesicle cargo through the recognition of short, linear amino-acid motifs in their cytoplasmic portions by vesicle coat proteins. For clathrin-coated vesicles, the motifs are recognized by clathrin adaptors. The AP2 adaptor complex (subunits α, β2, μ2 and σ2) recognizes both major endocytic motifs: YxxΦ motifs1 (where Φ can be F, I, L, M or V) and [ED]xxxL[LI] acidic dileucine motifs. Here we describe the binding of AP2 to the endocytic dileucine motif from CD4 (ref. 2). The major recognition events are the two leucine residues binding in hydrophobic pockets on σ2. The hydrophilic residue four residues upstream from the first leucine sits on a positively charged patch made from residues on the σ2 and α subunits. Mutations in key residues inhibit the binding of AP2 to ‘acidic dileucine’ motifs displayed in liposomes containing phosphatidylinositol-4,5-bisphosphate, but do not affect binding to YxxΦ motifs through μ2. In the ‘inactive’ AP2 core structure3 both motif-binding sites are blocked by different parts of the β2 subunit. To allow a dileucine motif to bind, the β2 amino terminus is displaced and becomes disordered; however, in this structure the YxxΦ-binding site on μ2 remains blocked.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Change history
18 December 2008
The AOP version of this paper carried an erroneous affiliation for D.J.O. This was corrected on 18 December 2008.
References
Owen, D. J. & Evans, P. R. A structural explanation for the recognition of tyrosine-based endocytotic signals. Science 282, 1327–1332 (1998)
Pitcher, C., Honing, S., Fingerhut, A., Bowers, K. & Marsh, M. Cluster of differentiation antigen 4 (CD4) endocytosis and adaptor complex binding require activation of the CD4 endocytosis signal by serine phosphorylation. Mol. Biol. Cell 10, 677–691 (1999)
Collins, B. M., McCoy, A. J., Kent, H. M., Evans, P. R. & Owen, D. J. Molecular architecture and functional model of the endocytic AP2 complex. Cell 109, 523–535 (2002)
Hurley, J. H., Lee, S. & Prag, G. Ubiquitin-binding domains. Biochem. J. 399, 361–372 (2006)
Pryor, P. R. et al. Molecular basis for the sorting of the SNARE VAMP7 into endocytic clathrin-coated vesicles by the ArfGAP Hrb. Cell 134, 817–827 (2008)
Miller, S. E., Collins, B. M., McCoy, A. J., Robinson, M. S. & Owen, D. J. A SNARE-adaptor interaction is a new mode of cargo recognition in clathrin-coated vesicles. Nature 450, 570–574 (2007)
Bonifacino, J. S. & Traub, L. M. Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 72, 395–447 (2003)
Shiba, T. et al. Structural basis for recognition of acidic-cluster dileucine sequence by GGA1. Nature 415, 937–941 (2002)
Misra, S., Puertollano, R., Kato, Y., Bonifacino, J. S. & Hurley, J. H. Structural basis for acidic-cluster-dileucine sorting-signal recognition by VHS domains. Nature 415, 933–937 (2002)
Chaudhuri, R., Lindwasser, O. W., Smith, W. J., Hurley, J. H. & Bonifacino, J. S. Downregulation of CD4 by human immunodeficiency virus type 1 Nef is dependent on clathrin and involves direct interaction of Nef with the AP2 clathrin adaptor. J. Virol. 81, 3877–3890 (2007)
Doray, B., Lee, I., Knisely, J., Bu, G. & Kornfeld, S. The γ/σ1 and α/σ2 hemicomplexes of clathrin adaptors AP-1 and AP-2 harbor the dileucine recognition site. Mol. Biol. Cell 18, 1887–1896 (2007)
Honing, S. et al. Phosphatidylinositol-(4,5)-bisphosphate regulates sorting signal recognition by the clathrin-associated adaptor complex AP2. Mol. Cell 18, 519–531 (2005)
Letourneur, F. & Klausner, R. D. A novel di-leucine motif and a tyrosine-based motif independently mediate lysosomal targeting and endocytosis of CD3 chains. Cell 69, 1143–1157 (1992)
Doray, B., Knisely, J. M., Wartman, L., Bu, G. & Kornfeld, S. Identification of acidic dileucine signals in LRP9 that interact with both GGAs and AP-1/AP-2. Traffic (in the press)
Huang, F., Jiang, X. & Sorkin, A. Tyrosine phosphorylation of the β2 subunit of clathrin adaptor complex AP-2 reveals the role of a di-leucine motif in the epidermal growth factor receptor trafficking. J. Biol. Chem. 278, 43411–43417 (2003)
Lee, I., Doray, B., Govero, J. & Kornfeld, S. Binding of cargo sorting signals to AP-1 enhances its association with ADP ribosylation factor 1-GTP. J. Cell Biol. 180, 467–472 (2008)
Mancias, J. D. & Goldberg, J. The transport signal on Sec22 for packaging into COPII-coated vesicles is a conformational epitope. Mol. Cell 26, 403–414 (2007)
Schwartz, T. & Blobel, G. Structural basis for the function of the β subunit of the eukaryotic signal recognition particle receptor. Cell 112, 793–803 (2003)
Bethune, J. et al. Coatomer, the coat protein of COPI transport vesicles, discriminates endoplasmic reticulum residents from p24 proteins. Mol. Cell. Biol. 26, 8011–8021 (2006)
McCoy, A. J., Grosse-Kunstleve, R. W., Storoni, L. C., Adams, P. D. & Read, R. J. PHASER crystallographic software. J. Appl. Crystallogr. 40, 658–674 (2007)
Leslie, A. G. The integration of macromolecular diffraction data. Acta Crystallogr. D 62, 48–57 (2006)
Evans, P. Scaling and assessment of data quality. Acta Crystallogr. D 62, 72–82 (2006)
Adams, P. D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D 58, 1948–1954 (2002)
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Painter, J. & Merritt, E. A. Optimal description of a protein structure in terms of multiple groups undergoing TLS motion. Acta Crystallogr. D 62, 439–450 (2006)
Ricotta, D., Conner, S. D., Schmid, S. L., von Figura, K. & Honing, S. Phosphorylation of the AP2 μ subunit by AAK1 mediates high affinity binding to membrane protein sorting signals. J. Cell Biol. 156, 791–795 (2002)
Jonsson, U. et al. Real-time biospecific interaction analysis using surface plasmon resonance and a sensor chip technology. Biotechniques 11, 620–627 (1991)
Acknowledgements
We thank the protein crystallography beamline staff at Diamond, especially E. Duke, K. McAuley and R. Flaig, for their support and assistance. D.J.O., B.T.K. and S.E.M. are funded by a Wellcome Trust Senior Research Fellowship to D.J.O. S.H. and K.S. are supported by grants from the Deutsche Forschungsgemeinschaft (SFB635 and SFB670).
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Information
This file contains Supplementary Figures S1-S9 with Legends and Supplementary Tables S1-S3. (PDF 2219 kb)
Supplementary Movie 1
Supplementary Movie 1 shows animation of the conformational change between the IP6-liganded, inactive conformation (2vgl) and the acidic dileucine motif bound conformations of the AP2 core. The two structures were superimposed on the σ2 subunit. Residues Y6 and F7 are highlighted in the closed structure (green balls) and the dileucine peptide is shown in gold. The two structures are alternated first in a "front" view, then from a "back" view, rotated by 180° (MOV 2055 kb)
Rights and permissions
About this article
Cite this article
Kelly, B., McCoy, A., Späte, K. et al. A structural explanation for the binding of endocytic dileucine motifs by the AP2 complex. Nature 456, 976–979 (2008). https://doi.org/10.1038/nature07422
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature07422
This article is cited by
-
Regulated secretion of mutant p53 negatively affects T lymphocytes in the tumor microenvironment
Oncogene (2024)
-
Vesicle trafficking and vesicle fusion: mechanisms, biological functions, and their implications for potential disease therapy
Molecular Biomedicine (2022)
-
The role of phosphatidylserine on the membrane in immunity and blood coagulation
Biomarker Research (2022)
-
Clathrin-associated AP-1 controls termination of STING signalling
Nature (2022)
-
Structural basis of an endocytic checkpoint that primes the AP2 clathrin adaptor for cargo internalization
Nature Structural & Molecular Biology (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.