Abstract
Diatoms are photosynthetic secondary endosymbionts found throughout marine and freshwater environments, and are believed to be responsible for around one-fifth of the primary productivity on Earth1,2. The genome sequence of the marine centric diatom Thalassiosira pseudonana was recently reported, revealing a wealth of information about diatom biology3,4,5. Here we report the complete genome sequence of the pennate diatom Phaeodactylum tricornutum and compare it with that of T. pseudonana to clarify evolutionary origins, functional significance and ubiquity of these features throughout diatoms. In spite of the fact that the pennate and centric lineages have only been diverging for 90 million years, their genome structures are dramatically different and a substantial fraction of genes (∼40%) are not shared by these representatives of the two lineages. Analysis of molecular divergence compared with yeasts and metazoans reveals rapid rates of gene diversification in diatoms. Contributing factors include selective gene family expansions, differential losses and gains of genes and introns, and differential mobilization of transposable elements. Most significantly, we document the presence of hundreds of genes from bacteria. More than 300 of these gene transfers are found in both diatoms, attesting to their ancient origins, and many are likely to provide novel possibilities for metabolite management and for perception of environmental signals. These findings go a long way towards explaining the incredible diversity and success of the diatoms in contemporary oceans.
Similar content being viewed by others
Main
The sequenced diatoms represent two of the major classes of diatoms—the bi/multipolar centrics (Mediophyceae), to which T. pseudonana belongs, and the pennates (Bacillariophyceae), to which P. tricornutum belongs (Supplementary Fig. 1). The earliest fossil deposit from centrics is 180 million years (Myr) old and that from pennates is 90 Myr old6,7. Although being the youngest, the pennates are by far the most diversified, and they are major components of both pelagic and benthic habitats7. They display a range of features, including their bilateral symmetry, that distinguish them from centric species. For example, they have amoeboid isogametes, by contrast with the motile sperm and oogamy observed in centric species; they are major biofoulers; they include toxic species; and they generally respond most strongly to mesoscale iron fertilization7,8. Furthermore, members of the raphid pennate clade can glide actively along surfaces.
The completed P. tricornutum genome is approximately 27.4 megabases (Mb) in size, which is slightly smaller than T. pseudonana (32.4 Mb), and P. tricornutum is predicted to contain fewer genes (10,402 as opposed to 11,776; Table 1, Supplementary Fig. 2). Gene identification and functional analysis was facilitated by the availability of more than 130,000 expressed sequence tags (ESTs) generated from cells grown under 16 different conditions. In total, 86% of gene predictions had EST support (Supplementary Table 1).
P. tricornutum shares 57% of its genes with T. pseudonana (see Supplementary Information for criteria used), of which 1,328 are absent from other sequenced eukaryotes (Table 1). The molecular divergence between the two diatoms was assessed by examining the percentage amino acid identity of 4,267 orthologous gene pairs (Table 2, Fig. 1). We found an average identity of 54.9% between diatom orthologues, in comparison with approximately 43% between the diatoms and a more distantly related heterokont, the non-photosynthetic oomycete Phytophthora sojae. This agrees with the predicted ancient separation (around 700 Myr ago) of these lineages9,10. The divergence between the two diatoms is similar to what is observed between Saccharomyces cerevisiae and the related yeast Kluyveromyces lactis, and about halfway between the Homo sapiens/Takifugu rubripes (pufferfish) divergence and the H. sapiens/Ciona intestinalis (sea squirt) divergence (Table 2, Fig. 1). The more rapid evolutionary rates of diatoms compared with other organismal groups (for example, the fish–mammal divergence probably occurred in the Proterozoic era earlier than 550 Myr ago11) is consistent with previous observations6,7. As has been found in the two yeasts12, no major conservation of gene order (synteny) could be detected between the two diatom genomes other than in a few examples of microclusters of up to eight genes (Supplementary Fig. 3). Furthermore, approximately two-thirds of intron positions are unique to each species (Supplementary Information). The widespread intron gain that has been reported in T. pseudonana13 was not found in P. tricornutum (Table 1), suggesting that it may be a recent event in the centric diatom.
Analysis of molecular divergence between the diatoms and other heterokonts, and comparison with selected hemiascomycetes and chordates. The diatom–oomycete pair displays the lowest amino acid identity (43.3%), in agreement with their proposed ancient separation, around 700 Myr ago10. The divergence between the pennate and centric diatom is similar to the fish–mammal divergence, which probably occurred in the Proterozoic era (550 Myr ago)11. The centric–pennate divergence, on the other hand, has been dated to at least 90 Myr ago7. In the figure, we represent the cumulative frequencies of amino acid identity across each set of potential orthologous pairs shown in Table 2.
Large-scale within-genome duplication events do not appear to have played a major role in driving the generation of diatom diversity (Supplementary Information), by contrast with what has been found in yeasts and metazoans14,15. The observed high levels of diatom species diversity must therefore have been generated by other mechanisms. Whereas intron gain may be one factor in centric diatoms, the dramatic expansion of diatom-specific copia-retrotransposable elements may have contributed in the P. tricornutum genome (Table 1, Supplementary Figs 2, 4). These elements also appear to have expanded in other pennate diatoms (Supplementary Information), so they may have been a significant driving force in the generation of pennate diatom diversity through transpositional duplications and subsequent genome fragmentation.
Diatoms, and heterokonts in general, are believed to be derived from a secondary endosymbiotic process that took place around one billion years ago between a red alga and a heterotrophic eukaryote16. Diatom chloroplast genomes have fewer genes than red algal chloroplast genomes, indicating that a number of chloroplast genes were transferred to the nucleus after secondary endosymbiosis, and a few more genes appear to be in the process of transfer in one diatom species or the other5. It is generally thought that the diatom mitochondrion originated in the host, and the mitochondrial gene complement is almost identical to that of haptophytes and cryptophytes, which are other algal phyla that may have originated from the same secondary endosymbiotic event. We used a phylogenomic approach to search for genes of red algal origin in the two diatoms and the two sequenced oomycetes, Phytophthora ramorum and Phytophthora sojae9, using Cyanidioschyzon merolae as reference red algal genome17. We classified 171 genes as being of red algal origin, on the basis of strong (>85%) bootstrap support for the red-alga-plus-heterokont clade (Supplementary Table 2). Of the 171 high-scoring genes, 108 were shared between the two diatoms and 74 (43%) were predicted to be plastid targeted. In addition, 11 of these genes were also present in oomycetes, as expected if the common ancestor of diatoms and oomycetes had a red algal plastid that was subsequently lost in the oomycetes9. The results of this survey support there being a red algal origin for the diatom plastid and many gene transfers from the red algal nucleus to the host nucleus before the former was lost.
A remarkably high number of P. tricornutum predicted genes appear to have been transferred between diatoms and bacteria (784; 7.5% of gene models). Specifically, by searching for orthologous genes in 739 prokaryotic genomes, followed by automated phylogenetic tree construction and manual curation, we confirmed that 587 putative P. tricornutum genes clustered with bacteria-only clades or formed a sister group to clades that included only bacterial genes (with or without other heterokonts). This finding indicates that horizontal gene transfer between bacteria and diatoms is pervasive and is much higher than has been found in other sequenced eukaryotes18,19. Of the 587 identified sequences, 42% are only found in P. tricornutum whereas 56% are present in both diatoms (Fig. 2a), attesting to their ancient origin. Only 73 sequences are shared between P. tricornutum and Phytophthora spp. (Fig. 2a, Supplementary Table 3), 59 of which are also present in T. pseudonana, suggesting that the vast majority of gene transfers occurred after the divergence of photosynthetic heterokonts and oomycetes.
a, Venn diagrams showing how many of the bacterial genes identified in P. tricornutum are also found in other heterokonts (left panel), and which bacterial classes are most related phylogenetically (right panel). In each case, the Venn diagrams indicate the number of trees in which the designated taxa occur within the same clade or in a sister clade of P. tricornutum. b, Breakdown of different bacterial groups that occur in the same clade or in a sister clade of P. tricornutum. ‘Unique’ denotes a gene found only in a particular bacterial class; ‘shared’ denotes a gene that is most similar to a gene of that specific bacterial class but that is also present in other bacterial groups. c, PhyML maximum likelihood tree (log likelihood ratio, -22,358.321320) as inferred from the amino acid sequences of the large subunit of NAD(P)H assimilatory nitrite reductase (NirB). The choice of protein evolution model was WAG with gamma-distributed rates (α = 0.80), as suggested by a ProtTest analysis of the alignment (see Supplementary Information for methods). Numbers above selected branches indicate maximum likelihood bootstrap support (100 replicates). Gene fusions and distinct open reading frames are indicated adjacent to the appropriate clades. In most cases, the large (NirB) and small (NirD) subunits of NAD(P)H assimilatory nitrite reductase are encoded by distinct open reading frames, but in diatoms and planctomycetes the nirD and nirB open reading frames have been fused to encode a single gene product. A total of 587 trees show evidence for prokaryotic origins of diatom genes and are available in Supplementary Information.
Many of the genes shared between diatoms and bacteria encode components that are likely to provide novel metabolic capacities, for example for organic carbon and nitrogen utilization20(xylanases and glucanases, prismane, carbon-nitrogen hydrolase, amidohydrolase), functioning of the diatom urea cycle3 (carbamoyl transferase, carbamate kinase, ornithine cyclodeaminase) and polyamine metabolism related to diatom cell wall silicification21 (S-adenosylmethionine (SAM)-dependent decarboxylases and methyltransferases). Others are likely to encode novel cell wall components, and to provide unorthodox mechanisms of DNA replication, repair and recombination for a eukaryotic cell (Supplementary Table 3).
Bacterial genes in diatoms do not appear to be derived from any one specific source, but from a range of origins including proteobacteria, cyanobacteria and archaea (Fig. 2a, b, Supplementary Table 3). Heterotrophic bacteria and cyanobacteria, especially diazotrophs and planctomycete bacteria, have been found in various close associations with diatoms22,23,24, which may explain the unprecedented levels of horizontal gene transfer events that appear to have occurred. In P. tricornutum, bacterial genes are distributed throughout the genome, although several clusters, as well as regions devoid of bacterial genes, can be observed (Supplementary Fig. 5). Some of these genes in diatoms share bacterial-specific gene fusions that support phylogenetic associations, such as assimilatory nitrite reductase B and D subunits; these are apparently of planctomycete origin (Fig. 2c).
Bacterial histidine-kinase-based phosphorelay two-component systems, which are involved in environmental sensing, also appear to be highly developed in diatoms. For example, P. tricornutum contains a wide range of two-component signalling proteins sometimes organized in novel domain associations (Fig. 3). One of these proteins bears the classical features of bacterial phytochrome photoreceptors, as previously noted in T. pseudonana3,4. Another domain combination present in both diatoms resembles aureochrome blue-light photoreceptors25, and P. tricornutum contains orthologues of LovHK and other light-dependent histidine kinases reported in bacteria26,27.
Domains likely to be involved in signalling are illustrated schematically and P. tricornutum protein identifiers are indicated on the left. Proteins corresponding to putative photoreceptors (aureochrome, phytochrome and LovHK) are indicated above (in grey, above the horizontal line). For further information about two-component systems, see Supplementary Information. Domain abbreviations are PAS, Per–Arnt–Sim; bZIP, basic region/leucine zipper; GAF, cGMP phosphodiesterase/adenylyl cyclase/FhlA; PHY, phytochrome; HK-N, histidine kinase N-terminal domain; RR, response regulator; LRR, leucine-rich repeat; LUX R, LuxR transcriptional activator; CHASE, cyclases/histidine kinases associated sensory extracellular.
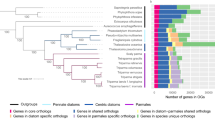
To identify additional novel features of the diatom gene repertoire, we compared the gene family content of the two diatoms with other eukaryotes (Fig. 4, Supplementary Figs 6, 7). Diatoms contain many species-specific multicopy gene families, as well as large numbers of species-specific single-copy genes (denoted orphans in Fig. 4a). The higher number of species-specific gene families in P. tricornutum may suggest that the more recent pennate diatoms possess more specialized functions, perhaps related to the heterogeneity of the benthic environments that they commonly inhabit. The centric diatom, by contrast, has retained more features found in other eukaryotes (Fig. 4b, Table 1), such as the flagellar apparatus28. We found a similar number of diatom-specific gene families (1,011) and eukaryotic gene families not found in diatoms (1,062), revealing that the rates of gene gain and gene loss are very similar and consistent with the high diversification rates observed in diatoms. We also found that diatom-specific genes are evolving faster than other genes in diatom genomes (Fig. 4c), providing a further explanation for the rapid diatom divergence rates6,7.
a, Venn diagram representation of shared/unique gene families in P. tricornutum, T. pseudonana, Viridiplantae (plants and green algae) and red algae, and other eukaryotes (that is, other chromalveolates and Opisthokonta (fungi and metazoa)). In addition to the total number of gene families specific to P. tricornutum and T. pseudonana, the number of families consisting of a single gene (denoted ‘orphans’) is also indicated. For example, of the 3,710 gene families that are only found in P. tricornutum, 3,423 consist of single-copy genes whereas 287 gene families have at least two members. b, Venn diagram of the distribution of P. tricornutum (left panel) and T. pseudonana (right panel) gene families with homology to proteins from the Viridiplantae & red algae, Opisthokonta and other chromalveolates (including the other diatom). The numbers outside the circles indicate the number of P. tricornutum (left panel) or T. pseudonana (right panel) gene families with no homology to the examined proteomes. c, Percentage amino acid identity plot of orthologues (based on reciprocal best hits) of different classes of diatom genes identified in a. Numbers in parentheses indicate the numbers of orthologues per class. ‘Diatom’ corresponds to genes only found in P. tricornutum and T. pseudonana (members of the 1,011 gene families shown in a), ‘core’ corresponds to genes present in all eukaryotic groups (members of the 1,666 gene families shown in a), and ‘all’ corresponds to all orthologous gene pairs in P. tricornutum and T. pseudonana.
Of the gene families found in the diatoms, some contain higher numbers of genes in comparison with other eukaryotes (Supplementary Table 4, Supplementary Fig. 7); for example, genes involved in polyamine metabolism are over-represented. The expansion of polyamine-related components is of interest in consideration of the role of long-chain polyamines in silica nanofabrication21. Of the eight predicted spermine/spermidine synthase-like genes in P. tricornutum, three encode potentially bifunctional enzymes bearing both an aminopropyltransferase domain and a SAM decarboxylase domain. Interestingly, the only other organisms containing such bifunctional proteins are T. pseudonana (four copies) and the bacteria Bdellovibrio bacteriovorus and Delftia acidovorans. Silaffins and silacidins are proteins/peptides believed to be involved in diatom silica formation21,29. P. tricornutum contains only one silaffin-like protein, and no homologues of silacidin. Frustulin genes, encoding proteins that form organic constituents of the biosilica cell wall but are not involved in silica formation, are present in large numbers and are highly expressed. Both diatoms contain a similar number of silicic acid transporters.
Other noteworthy diatom-specific expansions include histidine kinases (see above and Fig. 3), cyclins and heat-shock transcription factors. Cyclins are major regulators of the cell cycle in eukaryotes. In addition to members of each of the canonical families of cyclins, we found 10 and 42 diatom-specific cyclin genes in P. tricornutum and T. pseudonana, respectively. The dramatic expansion of this gene family may reflect the unusual characteristics of diatom life cycles due to the rigid nature of their cell walls, such as the control of cell size reduction, the activation of sexual reproduction at a critical size threshold, and life in rapidly changing and unpredictable environments7. Conversely, it may be significant that genes encoding RCC1 proteins (RCC, regulator of chromosome condensation), which are also involved in cell cycle control, have been expanded in both diatom genomes (Supplementary Table 4). For the putative heat-shock transcription factors, we found 69 copies in P. tricornutum and 89 copies in T. pseudonana4. These numbers represent almost 50% of the total number of transcription factors in the two sequenced diatoms. The significance of this expansion is unclear, but EST data indicates that the majority are expressed and that some are induced specifically in response to certain growth conditions (Supplementary Fig. 8).
In conclusion, through our comparative analyses we have revealed diverse origins of diatom genes. Diatom-specific genes may have arisen from genome rearrangements and subsequent domain recombinations due to the action of diatom-specific transposable elements, from selective gene family expansions/contractions and from intron gain/loss. It was previously shown that diatoms have retained genes from both partners of the secondary endosymbiosis3, thus bringing together primary metabolic processes such as photosynthetic carbon fixation and organic nitrogen production by means of the urea cycle in a single organism30. Our studies now suggest that genes acquired after secondary endosymbiosis by gene transfer from bacteria are pervasive in diatoms and represent at least 5% of their gene repertoires. This level of horizontal gene transfer is around one order of magnitude higher than has been found in other free-living eukaryotes, and is similar to the rates found between bacteria19. Although our analyses may be biased by the currently poor taxon sampling of whole genome sequences in eukaryotes (relative to that for prokaryotes), they are nonetheless supported by molecular phylogenies. We therefore propose that gene transfer from bacteria to diatoms, and perhaps vice versa, has been a common event in marine environments and has been a major driving force during diatom evolution. It has also brought together highly unorthodox combinations of genes permitting non-canonical management of carbon and nitrogen in primary metabolism and the sensing of external stimuli adapted to aquatic environments. The combination of mechanisms reported here may underlie the rapid diversification rates observed in diatoms and may explain why they have come to dominate contemporary marine ecosystems in a relatively short period of time.
Methods Summary
High-molecular-weight DNA was extracted from axenic cultures of P. tricornutum accession Pt1 8.6 (deposited as CCMP2561 in the Provasoli–Guillard National Center for Culture of Marine Phytoplankton) and used to construct replicate libraries containing inserts of 2–3 kb, 6–8 kb and 35–40 kb. Using the Joint Genome Institute (JGI) JAZZ assembler, approximately 556,000 reads involving 564 Mb of sequence were trimmed, filtered for short reads and assembled. All low-quality areas and gaps were identified and converted into targets for manual finishing. The draft genome sequence of T. pseudonana3 was finished in a similar way. Both diatom genomes were annotated using the JGI annotation pipeline, which combines several gene prediction, annotation and analysis tools. Complementary DNA libraries were constructed from messenger RNA extracted from P. tricornutum cultures grown under 16 different conditions. More than 130,000 ESTs were generated. Full information about all methods used for the analyses reported here is available in Supplementary Information.
Accession codes
Primary accessions
DDBJ/GenBank/EMBL
EMBL/GenBank/DDBJ
GenBank/EMBL/DDBJ
Data deposits
Assemblies and annotations of the P. tricornutum and T. pseudonana genomes are available through the JGI Genome Portal at http://www.jgi.doe.gov/phaeodactylum and http://www.jgi.doe.gov/thalassiosira. Genome assemblies together with predicted gene models and annotations have been deposited at DDBJ, EMBL and GenBank under the project accessions ABQD00000000 and AAFD00000000, respectively. The versions described in this paper are the first version, ABQD01000000, for P. tricornutum, which includes complete chromosomes 3 and 11 (CP001142 and CP001141), and the second version, AAFD02000000, for T. pseudonana, also including complete chromosomes 7 and 18 (CP001160 and CP001159). P. tricornutum EST expression profiles can be found at http://www.biologie.ens.fr/diatomics/EST3, which also provides links to gene models on the JGI genome browser. ESTs have been deposited at NCBI dbEST with GenBank accession numbers CD374840-CD384835, BI306757-BI307753, CT868744-CT950687 and CU695349-CU740080.
References
Falkowski, P. G., Barber, R. T. & Smetacek, V. Biogeochemical controls and feedbacks on ocean primary production. Science 281, 200–206 (1998)
Field, C. B., Behrenfeld, M. J., Randerson, J. T. & Falkowski, P. Primary production of the biosphere: integrating terrestrial and oceanic components. Science 281, 237–240 (1998)
Armbrust, E. V. et al. The genome of the diatom Thalassiosira pseudonana: ecology, evolution, and metabolism. Science 306, 79–86 (2004)
Montsant, A. et al. Identification and comparative genomic analysis of signaling and regulatory components in the diatom Thalassiosira pseudonana . J. Phycol. 43, 585–603 (2007)
Oudot-Le Secq, M.-P. et al. Chloroplast genomes of the diatoms Phaeodactylum tricornutum and Thalassiosira pseudonana: comparison with other plastid genomes of the red lineage. Mol. Gen. Genom. 277, 427–439 (2007)
Sims, P. A., Mann, D. G. & Medlin, L. K. Evolution of the diatoms: insights from fossil, biological and molecular data. Phycologia 45, 361–402 (2006)
Kooistra, W. H. C. F., Gersonde, R., Medlin, L. K. & Mann, D. G. in Evolution of Primary Producers in the Sea (eds Falkowski, P. G. & Knoll, A. H.) 207–249 (Academic Press, 2007)
de Baar, H. J. W. et al. Synthesis of iron fertilization experiments: From the Iron Age in the Age of Enlightenment. J. Geophys. Res. 110 doi: 10.1029/2004JC002601 (2005)
Tyler, B. M. et al. Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science 313, 1261–1266 (2006)
Yoon, H. S., Hackett, J. D., Ciniglia, C., Pinto, G. & Bhattacharya, D. A molecular timeline for the origin of photosynthetic eukaryotes. Mol. Biol. Evol. 21, 809–818 (2004)
Kumar, S. & Hedges, S. B. A molecular timescale for vertebrate evolution. Nature 392, 917–920 (1998)
Dujon, B. Yeasts illustrate the molecular mechanisms of eukaryotic genome evolution. Trends Genet. 22, 375–387 (2006)
Roy, S. W. & Penny, D. A very high fraction of unique intron positions in the intron-rich diatom Thalassiosira pseudonana indicates widespread intron gain. Mol. Biol. Evol. 24, 1447–1457 (2007)
Scannell, D. R., Byrne, K. P., Gordon, J. L., Wong, S. & Wolfe, K. H. Multiple rounds of speciation associated with reciprocal gene loss in polyploid yeasts. Nature 440, 341–345 (2006)
Semon, M. & Wolfe, K. H. Reciprocal gene loss between Tetraodon and zebrafish after whole genome duplication in their ancestor. Trends Genet. 23, 108–112 (2007)
Bhattacharya, D., Archibald, J. M., Weber, A. P. & Reyes-Prieto, A. How do endosymbionts become organelles? Understanding early events in plastid evolution. Bioessays 29, 1239–1246 (2007)
Matsuzaki, M. et al. Genome sequence of the ultrasmall unicellular red alga Cyanidioschyzon merolae 10D. Nature 428, 653–657 (2004)
Martens, C., Vandepoele, K. & Van de Peer, Y. Whole-genome analysis reveals molecular innovations and evolutionary transitions in chromalveolate species. Proc. Natl Acad. Sci. USA 105, 3427–3432 (2008)
Keeling, P. J. & Palmer, J. D. Horizontal gene transfer in eukaryotic evolution. Nature Rev. Genet. 9, 605–618 (2008)
Kroth, P. G. et al. A model for carbohydrate metabolism in the diatom Phaeodactylum tricornutum deduced from whole genome analysis and comparative genomic analyses with Thalassiosira pseudonana and other photoautotrophs. PLoS One 3, e1426 (2008)
Kröger, N. Prescribing diatom morphology: toward genetic engineering of biological nanomaterials. Curr. Opin. Chem. Biol. 11, 662–669 (2007)
Carpenter, E. J. & Janson, S. Intracellular cyanobacterial symbionts in the marine diatom Climacodium frauenfeldianum (Bacillariophyceae). J. Phycol. 36, 540–544 (2000)
Schmid, A.-M. M. Endobacteria in the diatom Pinnularia (Bacillariophyceae). I. “Scattered ct-nucleoids” explained: DAPI-DNA complexes stem from exoplastidial bacteria boring into the chloroplasts. J. Phycol. 39, 122–138 (2003)
Zehr, J. P., Carpenter, E. J. & Villareal, T. A. New perspectives on nitrogen-fixing microorganisms in tropical and subtropical oceans. Trends Microbiol. 8, 68–73 (2000)
Takahashi, F. et al. AUREOCHROME, a photoreceptor required for photomorphogenesis in stramenopiles. Proc. Natl Acad. Sci. USA 104, 19625–19630 (2007)
Purcell, E. B., Siegal-Gaskins, D., Rawling, D. C., Fiebig, A. & Crosson, S. A photosensory two-component system regulates bacterial cell attachment. Proc. Natl Acad. Sci. USA 104, 18241–18246 (2007)
Swartz, T. E. et al. Blue-light-activated histidine kinases: two-component sensors in bacteria. Science 317, 1090–1093 (2007)
Merchant, S. S. et al. The Chlamydomonas genome reveals the evolution of key animal and plant functions. Science 318, 245–250 (2007)
Sumper, M. & Brunner, E. Silica biomineralization in diatoms: the model organism Thalassiosira pseudonana . ChemBioChem 9, 1187–1194 (2008)
Allen, A. E., Vardi, A. & Bowler, C. An ecological and evolutionary context for integrated nitrogen metabolism and related signaling pathways in marine diatoms. Curr. Opin. Plant Biol. 9, 264–273 (2006)
Acknowledgements
Diatom genome sequencing at the JGI (USA) was performed under the auspices of the US Department of Energy’s Office of Science, Biological and Environmental Research Program, and by the University of California, Lawrence Berkeley National Laboratory, under contract no. DE-AC02-05CH11231, Lawrence Livermore National Laboratory under contract no. DE-AC52-07NA27344 and Los Alamos National Laboratory under contract no. DE-AC02-06NA25396. P. tricornutum ESTs were generated at Genoscope (France). Funding for this work was also obtained from the EU-funded FP6 Diatomics project (LSHG-CT-2004-512035), the EU-FP6 Marine Genomics Network of Excellence (GOCE-CT-2004-505403), an ATIP ‘Blanche’ grant from the CNRS (France) and the Agence Nationale de la Recherche (France). We are grateful to M. Muffato and H.-R. Crollius for the analysis reported in Supplementary Fig. 3a.
Author Contributions C.B. coordinated Phaeodactylum genome annotation and manuscript preparation. E.V.A. coordinated Thalassiosira genome annotation. D.S.R. and I.V.G. coordinated diatom genome sequencing and analysis at JGI. J.W. coordinated EST sequencing at Genoscope. A.E.A., J.H.B., J.G., K.J., A.K., U.M., C.M., F.M., R.P.O., E.R., A.S. and K.V. made equivalent and substantial contributions to the data presented, and should be considered joint second authors. B.B., A.G., M.H., M.K., T.M., K.V. and F.V. also made significant contributions. A.E.A., E.V.A., B.R.G., Y.V.d.P. and I.V.G. assisted in data interpretation and manuscript preparation. Other authors contributed as members of the Phaeodactylum genome sequencing consortium.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Information
This file contains Supplementary Figures 1-8 with Legends, Supplementary Tables 1-2, 4 and 6-8 (see separate files for Tables 3 and 5), Supplementary Methods and Notes and Supplementary References (PDF 2390 kb)
Supplementary Table 3
This file contains Supplementary Table 3 (legend in nature07410-s1.pdf) (PDF 111 kb)
Supplementary Table 5
This file contains Supplementary Table 5 (legend in nature07410-s1.pdf) (PDF 117 kb)
Supplementary Information
This file contains the 587 phylogenetic trees of P. tricornutem genes of proposed bacterial origin (PDF 4747 kb)
Rights and permissions
This article is distributed under the terms of the Creative Commons Attribution-Non-Commercial-Share Alike licence (http://creativecommons.org/licenses/by-nc-sa/3.0/), which permits distribution, and reproduction in any medium, provided the original author and source are credited. This licence does not permit commercial exploitation, and derivative works must be licensed under the same or similar licence.
About this article
Cite this article
Bowler, C., Allen, A., Badger, J. et al. The Phaeodactylum genome reveals the evolutionary history of diatom genomes. Nature 456, 239–244 (2008). https://doi.org/10.1038/nature07410
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature07410
This article is cited by
-
A key gene, violaxanthin de-epoxidase-like 1, enhances fucoxanthin accumulation in Phaeodactylum tricornutum
Biotechnology for Biofuels and Bioproducts (2024)
-
Transcriptomic and metabolomic analyses revealed regulation mechanism of mixotrophic Cylindrotheca sp. glycerol utilization and biomass promotion
Biotechnology for Biofuels and Bioproducts (2023)
-
The genomes of Vischeria oleaginous microalgae shed light on the molecular basis of hyper-accumulation of lipids
BMC Biology (2023)
-
Genome analysis of Parmales, the sister group of diatoms, reveals the evolutionary specialization of diatoms from phago-mixotrophs to photoautotrophs
Communications Biology (2023)
-
PhaeoEpiView: an epigenome browser of the newly assembled genome of the model diatom Phaeodactylum tricornutum
Scientific Reports (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.