Abstract
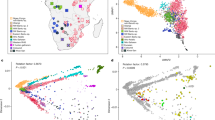
Understanding the genetic structure of human populations is of fundamental interest to medical, forensic and anthropological sciences. Advances in high-throughput genotyping technology have markedly improved our understanding of global patterns of human genetic variation and suggest the potential to use large samples to uncover variation among closely spaced populations1,2,3,4,5. Here we characterize genetic variation in a sample of 3,000 European individuals genotyped at over half a million variable DNA sites in the human genome. Despite low average levels of genetic differentiation among Europeans, we find a close correspondence between genetic and geographic distances; indeed, a geographical map of Europe arises naturally as an efficient two-dimensional summary of genetic variation in Europeans. The results emphasize that when mapping the genetic basis of a disease phenotype, spurious associations can arise if genetic structure is not properly accounted for. In addition, the results are relevant to the prospects of genetic ancestry testing6; an individual’s DNA can be used to infer their geographic origin with surprising accuracy—often to within a few hundred kilometres.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Jakobsson, M. et al. Genotype, haplotype and copy-number variation in worldwide human populations. Nature 451, 998–1003 (2008)
Li, J. Z. et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science 319, 1100–1104 (2008)
Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature 447, 661–678 (2007)
Tian, C. et al. Analysis and application of European genetic substructure using 300K SNP information. PLoS Genet. 4, e4 (2008)
Price, A. L. et al. Discerning the ancestry of European Americans in genetic association studies. PLoS Genet. 4, e236 (2008)
Shriver, M. D. & Kittles, R. A. Genetic ancestry and the search for personalized genetic histories. Nature Rev. Genet. 5, 611–618 (2004)
Nelson, M. R. et al. The Population Reference Sample (POPRES): a resource for population, disease, and pharmacological genetics research. Am. J. Hum. Genet. (in the press)
Patterson, N., Price, A. & Reich, D. Population structure and eigenanalysis. PLoS Genet. 2, e190 (2006)
Novembre, J. & Stephens, M. Interpreting principal component analyses of spatial population genetic variation. Nature Genet. 40, 646–649 (2008)
Menozzi, P., Piazza, A. & Cavalli-Sforza, L. Synthetic maps of human gene frequencies in Europeans. Science 201, 786–792 (1978)
Campbell, C. D. et al. Demonstrating stratification in a European American population. Nature Genet. 37, 868–872 (2005)
Price, A. L. et al. Principal components analysis corrects for stratification in genome-wide association studies. Nature Genet. 38, 904–909 (2006)
McCarthy, M. I. et al. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nature Rev. Genet. 9, 356–369 (2008)
Zhu, X., Zhang, S., Zhao, H. & Cooper, R. S. Association mapping, using a mixture model for complex traits. Genet. Epidemiol. 23, 181–196 (2002)
Weedon, M. N. et al. Genome-wide association analysis identifies 20 loci that influence adult height. Nature Genet. 40, 575–583 (2008)
Lettre, G. et al. Identification of ten loci associated with height highlights new biological pathways in human growth. Nature Genet. 40, 584–591 (2008)
Cavalli-Sforza, L. L., Menozzi, P. & Piazza, A. The History and Geography of Human Genes 292 (Princeton Univ. Press, 1994)
Bauchet, M. et al. Measuring European population stratification with microarray genotype data. Am. J. Hum. Genet. 80, 948–956 (2007)
Weir, B. S. & Cockerham, C. C. Estimating F-statistics for the analysis of population structure. Evolution 38, 1358–1370 (1984)
Eberle, M. A. & Kruglyak, L. An analysis of strategies for discovery of single nucleotide polymorphisms. Genet. Epidemiol. 19, S29–S35 (2000)
Clark, A. G., Hubisz, M. J., Bustamante, C. D., Williamson, S. H. & Nielsen, R. Ascertainment bias in studies of human genome-wide polymorphism. Genome Res. 15, 1496–1502 (2005)
Slatkin, M. Rare alleles as indicators of gene flow. Evolution 39, 53–65 (1985)
Falush, D., Stephens, M. & Pritchard, J. K. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164, 1567–1587 (2003)
Tang, H., Coram, M., Wang, P., Zhu, X. & Risch, N. Reconstructing genetic ancestry blocks in admixed individuals. Am. J. Hum. Genet. 79, 1–12 (2006)
Hellenthal, G., Auton, A. & Falush, D. Inferring human colonization history using a copying model. PLoS Genet. 4, e1000078 (2008)
Kooner, J. et al. Genome-wide scan identifies variation in MLXIPL associated with plasma triglycerides. Nature Genet. 40, 149–151 (2008)
Firmann, M. et al. The CoLaus study: A population-based study to investigate the epidemiology and genetic determinants of cardiovascular risk factors and metabolic syndrome. BMC Cardiovasc. Dis. 8, 6 (2008)
Purcell, S. et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007)
Acknowledgements
We thank J. Kooner and J. Chambers of the LOLIPOP study and G. Waeber, P. Vollenweider, D. Waterworth, J. S. Beckmann, M. Bochud and V. Mooser of the CoLaus study for providing access to their collections. Financial support was provided by the Giorgi-Cavaglieri Foundation (S.B.), the Swiss National Science Foundation (S.B.), US National Science Foundation Postdoctoral Fellowship in Bioinformatics (J.N.), US National Institutes of Health (M.S., C.D.B.) and GlaxoSmithKline (M.R.N.).
Author Contributions M.R.N. coordinated sample collection and genotyping. K.S.K., A.I., J.N. and A.R.B. performed quality control and prepared genotypic and demographic data for further analyses. C.B., M.S., M.R.N., S.B., J.N., T.J., K.B., Z.K., A.R.B. and A.A. all contributed to the design of analyses. J.N., S.B., T.J., K.B. and Z.K. performed PCA analyses. M.S. and J.N. designed and performed assignment-based analyses. T.J. and J.N. performed genome-wide association simulations. J.N., C.B., M.S., M.R.N. and A.A. wrote the paper. All authors discussed the results and commented on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Supplementary information
Supplementary Information
This file contains Supplementary Notes, Supplementary Figures 1-6 with legends, and Supplementary Tables 1-5. (PDF 715 kb)
PowerPoint slides
Rights and permissions
About this article
Cite this article
Novembre, J., Johnson, T., Bryc, K. et al. Genes mirror geography within Europe. Nature 456, 98–101 (2008). https://doi.org/10.1038/nature07331
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature07331
This article is cited by
-
Differentiated adaptative genetic architecture and language-related demographical history in South China inferred from 619 genomes from 56 populations
BMC Biology (2024)
-
GrassCaré: Visualizing the Grassmannian on the Poincaré Disk
SN Computer Science (2024)
-
The genetic legacy of the expansion of Bantu-speaking peoples in Africa
Nature (2024)
-
Genetic risk scores: are they important for diabetes management? results from multiple cross-sectional studies
Diabetology & Metabolic Syndrome (2023)
-
How can gender be identified from heart rate data? Evaluation using ALLSTAR heart rate variability big data analysis
BMC Research Notes (2023)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.