Abstract
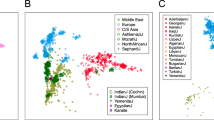
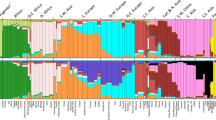
Genome-wide patterns of variation across individuals provide a powerful source of data for uncovering the history of migration, range expansion, and adaptation of the human species. However, high-resolution surveys of variation in genotype, haplotype and copy number have generally focused on a small number of population groups1,2,3. Here we report the analysis of high-quality genotypes at 525,910 single-nucleotide polymorphisms (SNPs) and 396 copy-number-variable loci in a worldwide sample of 29 populations. Analysis of SNP genotypes yields strongly supported fine-scale inferences about population structure. Increasing linkage disequilibrium is observed with increasing geographic distance from Africa, as expected under a serial founder effect for the out-of-Africa spread of human populations. New approaches for haplotype analysis produce inferences about population structure that complement results based on unphased SNPs. Despite a difference from SNPs in the frequency spectrum of the copy-number variants (CNVs) detected—including a comparatively large number of CNVs in previously unexamined populations from Oceania and the Americas—the global distribution of CNVs largely accords with population structure analyses for SNP data sets of similar size. Our results produce new inferences about inter-population variation, support the utility of CNVs in human population-genetic research, and serve as a genomic resource for human-genetic studies in diverse worldwide populations.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Accession codes
Primary accessions
Gene Expression Omnibus
Data deposits
The array data described in this paper are deposited in the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession number GSE10331.
References
The International Haplotype Map Consortium. A haplotype map of the human genome. Nature 437, 1299–1320 (2005)
Hinds, D. A. et al. Whole-genome patterns of common DNA variation in three human populations. Science 307, 1072–1079 (2005)
Redon, R. et al. Global variation in copy number in the human genome. Nature 444, 444–454 (2006)
Cann, H. M. et al. A human genome diversity cell line panel. Science 296, 261–262 (2002)
Kalinowski, S. T. Counting alleles with rarefaction: private alleles and hierarchical sampling designs. Conserv. Genet. 5, 539–543 (2004)
Falush, D., Stephens, M. & Pritchard, J. K. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164, 1567–1587 (2003)
Bastos-Rodrigues, L., Pimenta, J. R. & Pena, S. D. J. The genetic structure of human populations studied through short insertion–deletion polymorphisms. Ann. Hum. Genet. 70, 658–665 (2006)
Rosenberg, N. A. et al. Clines, clusters, and the effect of study design on the inference of human population structure. PLoS Genet. 1, e70 (2005)
Rosenberg, N. A. et al. Genetic structure of human populations. Science 298, 2381–2385 (2002)
Lawson Handley, L. J., Manica, A., Goudet, J. & Balloux, F. Going the distance: human population genetics in a clinal world. Trends Genet. 23, 432–439 (2007)
Ramachandran, S. et al. Support from the relationship of genetic and geographic distance in human populations for a serial founder effect originating in Africa. Proc. Natl Acad. Sci. USA 102, 15942–15947 (2005)
Sabatti, C. & Risch, N. Homozygosity and linkage disequilibrium. Genetics 160, 1707–1719 (2002)
Conrad, D. F. et al. A worldwide survey of haplotype variation and linkage disequilibrium in the human genome. Nature Genet. 38, 1251–1260 (2006)
Gabriel, S. B. et al. The structure of haplotype blocks in the human genome. Science 296, 2225–2229 (2002)
Reich, D. E. et al. Linkage disequilibrium in the human genome. Nature 411, 199–204 (2001)
Tishkoff, S. A. & Kidd, K. K. Implications of biogeography of human populations for ‘race’ and medicine. Nature Genet. 36, S21–S27 (2004)
McVean, G. A. T. A genealogical interpretation of linkage disequilibrium. Genetics 162, 987–991 (2002)
Bersaglieri, T. et al. Genetic signatures of strong recent positive selection at the lactase gene. Am. J. Hum. Genet. 74, 1111–1120 (2004)
Tishkoff, S. A. et al. Convergent adaptation of human lactase persistence in Africa and Europe. Nature Genet. 39, 31–40 (2007)
Wang, K. et al. PennCNV: an integrated hidden Markov model designed for high-resolution copy number variation detection in whole-genome SNP genotyping data. Genome Res. 17, 1665–1674 (2007)
Wong, K. K. et al. A comprehensive analysis of common copy-number variations in the human genome. Am. J. Hum. Genet. 80, 91–104 (2007)
Locke, D. P. et al. Linkage disequilibrium and heritability of copy-number polymorphisms within duplicated regions of the human genome. Am. J. Hum. Genet. 79, 275–290 (2006)
Sharp, A. J. et al. Segmental duplications and copy-number variation in the human genome. Am. J. Hum. Genet. 77, 78–88 (2005)
Scherer, S. W. et al. Challenges and standards in integrating surveys of structural variation. Nature Genet. 39, S7–S15 (2007)
Servin, B. & Stephens, M. Imputation-based analysis of association studies: candidate regions and quantitative traits. PLoS Genet. 3, e114 (2007)
Need, A. C. & Goldstein, D. B. Genome-wide tagging for everyone. Nature Genet. 38, 1227–1228 (2006)
Eberle, M. A., Rieder, M. J., Kruglyak, L. & Nickerson, D. A. Allele frequency matching between SNPs reveals an excess of linkage disequilibrium in genic regions of the human genome. PLoS Genet. 2, e142 (2006)
Scheet, P. & Stephens, M. A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am. J. Hum. Genet. 78, 629–644 (2006)
Jakobsson, M. & Rosenberg, N. A. CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23, 1801–1806 (2007)
Zhang, J., Feuk, L., Duggan, G. E., Khaja, R. & Scherer, S. W. Development of bioinformatics resources for display and analysis of copy number and other structural variants in the human genome. Cytogenet. Genome Res. 115, 205–214 (2006)
Acknowledgements
We thank the Biological Resource Center at the Fondation Jean Dausset – CEPH for preparing HGDP–CEPH diversity panel DNA samples, and S. Chanock and A. Hutchinson for assistance with the DNAs. This work was supported in part by NIH grants, by a postdoctoral fellowship from the University of Michigan Center for Genetics in Health and Medicine, by grants from the Alfred P. Sloan Foundation and the Burroughs Wellcome Fund, by the National Center for Minority Health and Health Disparities, and by the Intramural Program of the National Institute on Aging. The study used the Biowulf Linux cluster at the National Institutes of Health (http://biowulf.nih.gov).
Author Contributions N.A.R. and A.B.S. wish to be regarded as joint last authors.
Author information
Authors and Affiliations
Corresponding authors
Supplementary information
Supplementary Information
This file contains extensive Supplementary Information with Supplementary Notes, Supplementary Data, Supplementary Tables S1-S17, Supplementary Figures S1-S30 with Legends and additional references. (PDF 10195 kb)
Rights and permissions
About this article
Cite this article
Jakobsson, M., Scholz, S., Scheet, P. et al. Genotype, haplotype and copy-number variation in worldwide human populations. Nature 451, 998–1003 (2008). https://doi.org/10.1038/nature06742
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature06742
This article is cited by
-
The Allen Ancient DNA Resource (AADR) a curated compendium of ancient human genomes
Scientific Data (2024)
-
Hybrid autoencoder with orthogonal latent space for robust population structure inference
Scientific Reports (2023)
-
An integrative framework and recommendations for the study of DNA methylation in the context of race and ethnicity
Discover Social Science and Health (2023)
-
Comprehensive genome-wide association study of different forms of hernia identifies more than 80 associated loci
Nature Communications (2022)
-
Association between TAP gene polymorphisms and tuberculosis susceptibility in a Han Chinese population in Guangdong
Molecular Genetics and Genomics (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.