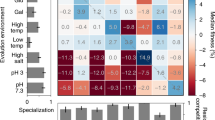
Abstract
How gene duplication and divergence contribute to genetic novelty and adaptation has been of intense interest, but experimental evidence has been limited. The genetic switch controlling the yeast galactose use pathway includes two paralogous genes in Saccharomyces cerevisiae that encode a co-inducer (GAL3) and a galactokinase (GAL1). These paralogues arose from a single bifunctional ancestral gene as is still present in Kluyveromyces lactis. To determine which evolutionary processes shaped the evolution of the two paralogues, here we assess the effects of precise replacement of coding and non-coding sequences on organismal fitness. We suggest that duplication of the ancestral bifunctional gene allowed for the resolution of an adaptive conflict between the transcriptional regulation of the two gene functions. After duplication, previously disfavoured binding site configurations evolved that divided the regulation of the ancestral gene into two specialized genes, one of which ultimately became one of the most tightly regulated genes in the genome.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Ohno, S. Evolution by Gene Duplication (Springer, New York, 1970)
Taylor, J. S. & Raes, J. Duplication and divergence: the evolution of new genes and old ideas. Annu. Rev. Genet. 38, 615–643 (2004)
Irwin, D. M., Prager, E. M. & Wilson, A. C. Evolutionary genetics of ruminant lysozymes. Anim. Genet. 23, 193–202 (1992)
Yokoyama, S. Molecular evolution of color vision in vertebrates. Gene 300, 69–78 (2002)
Zhang, J., Zhang, Y. P. & Rosenberg, H. F. Adaptive evolution of a duplicated pancreatic ribonuclease gene in a leaf-eating monkey. Nature Genet. 30, 411–415 (2002)
Prince, V. E. & Pickett, F. B. Splitting pairs: the diverging fates of duplicated genes. Nature Rev. Genet. 3, 827–837 (2002)
van Hoof, A. Conserved functions of yeast genes support the duplication, degeneration and complementation model for gene duplication. Genetics 171, 1455–1461 (2005)
Scannell, D. R., Byrne, K. P., Gordon, J. L., Wong, S. & Wolfe, K. H. Multiple rounds of speciation associated with reciprocal gene loss in polyploid yeasts. Nature 440, 341–345 (2006)
Schilke, B. et al. Evolution of mitochondrial chaperones utilized in Fe–S cluster biogenesis. Curr. Biol. 16, 1660–1665 (2006)
Piatigorsky, J. Gene Sharing and Evolution: The Diversity of Protein Functions (Harvard Univ. Press, Cambridge, Massachusetts, 2007)
Force, A. et al. Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151, 1531–1545 (1999)
Tumpel, S., Cambronero, F., Wiedemann, L. M. & Krumlauf, R. Evolution of cis elements in the differential expression of two Hoxa2 coparalogous genes in pufferfish (Takifugu rubripes). Proc. Natl Acad. Sci. USA 103, 5419–5424 (2006)
Piatigorsky, J. & Wistow, G. The recruitment of crystallins: new functions precede gene duplication. Science 252, 1078–1079 (1991)
Hughes, A. L. The evolution of functionally novel proteins after gene duplication. Proc. Biol. Sci. 256, 119–124 (1994)
Hughes, A. L. Adaptive Evolution of Genes and Genomes (Oxford Univ. Press, Oxford, 1999)
Johnston, M. A model fungal gene regulatory mechanism: the GAL genes of Saccharomyces cerevisiae . Microbiol. Rev. 51, 458–476 (1987)
Bhat, P. J. & Murthy, T. V. Transcriptional control of the GAL/MEL regulon of yeast Saccharomyces cerevisiae: mechanism of galactose-mediated signal transduction. Mol. Microbiol. 40, 1059–1066 (2001)
Ptashne, M. & Gann, A. Genes and Signals (Cold Spring Harbor Laboratory Press, Woodbury, New York, 2001)
Wolfe, K. H. & Shields, D. C. Molecular evidence for an ancient duplication of the entire yeast genome. Nature 387, 708–713 (1997)
Dietrich, F. S. et al. The Ashbya gossypii genome as a tool for mapping the ancient Saccharomyces cerevisiae genome. Science 304, 304–307 (2004)
Kellis, M., Birren, B. W. & Lander, E. S. Proof and evolutionary analysis of ancient genome duplication in the yeast Saccharomyces cerevisiae . Nature 428, 617–624 (2004)
Hittinger, C. T., Rokas, A. & Carroll, S. B. Parallel inactivation of multiple GAL pathway genes and ecological diversification in yeasts. Proc. Natl Acad. Sci. USA 101, 14144–14149 (2004)
Scannell, D. R. et al. Independent sorting-out of thousands of duplicated gene pairs in two yeast species descended from a whole-genome duplication. Proc. Natl Acad. Sci. USA 104, 8397–8402 (2007)
Johnston, M. & Davis, R. W. Sequences that regulate the divergent GAL1–GAL10 promoter in Saccharomyces cerevisiae. . Mol. Cell. Biol. 4, 1440–1448 (1984)
West, R. W., Yocum, R. R. & Ptashne, M. Saccharomyces cerevisiae GAL1–GAL10 divergent promoter region: location and function of the upstream activating sequence UASG . Mol. Cell. Biol. 4, 2467–2478 (1984)
Bajwa, W., Torchia, T. E. & Hopper, J. E. Yeast regulatory gene GAL3: carbon regulation; UASGal elements in common with GAL1, GAL2, GAL7, GAL10, GAL80, and MEL1; encoded protein strikingly similar to yeast and Escherichia coli galactokinases. Mol. Cell. Biol. 8, 3439–3447 (1988)
Zenke, F. T. et al. Activation of Gal4p by galactose-dependent interaction of galactokinase and Gal80p. Science 272, 1662–1665 (1996)
Yano, K. & Fukasawa, T. Galactose-dependent reversible interaction of Gal3p with Gal80p in the induction pathway of Gal4p-activated genes of Saccharomyces cerevisiae . Proc. Natl Acad. Sci. USA 94, 1721–1726 (1997)
Platt, A. & Reece, R. J. The yeast galactose genetic switch is mediated by the formation of a Gal4p-Gal80p-Gal3p complex. EMBO J. 17, 4086–4091 (1998)
Peng, G. & Hopper, J. E. Gene activation by interaction of an inhibitor with a cytoplasmic signaling protein. Proc. Natl Acad. Sci. USA 99, 8548–8553 (2002)
Meyer, J., Walker-Jonah, A. & Hollenberg, C. P. Galactokinase encoded by GAL1 is a bifunctional protein required for induction of the GAL genes in Kluyveromyces lactis and is able to suppress the gal3 phenotype in Saccharomyces cerevisiae . Mol. Cell. Biol. 11, 5454–5461 (1991)
Rubio-Texeira, M. A comparative analysis of the GAL genetic switch between not-so-distant cousins: Saccharomyces cerevisiae versus Kluyveromyces lactis . FEMS Yeast Res. 5, 1115–1128 (2005)
Webster, T. D. & Dickson, R. C. The organization and transcription of the galactose gene cluster of Kluyveromyces lactis . Nucleic Acids Res. 16, 8011–8028 (1988)
Thoden, J. B., Sellick, C. A., Timson, D. J., Reece, R. J. & Holden, H. M. Molecular structure of Saccharomyces cerevisiae Gal1p, a bifunctional galactokinase and transcriptional inducer. J. Biol. Chem. 280, 36905–36911 (2005)
Platt, A., Ross, H. C., Hankin, S. & Reece, R. J. The insertion of two amino acids into a transcriptional inducer converts it into a galactokinase. Proc. Natl Acad. Sci. USA 97, 3154–3159 (2000)
Weinreich, D. M., Watson, R. A. & Chao, L. Perspective: sign epistasis and genetic constraint on evolutionary trajectories. Evol. Int. J. Org. Evol. 59, 1165–1174 (2005)
Kellis, M., Patterson, N., Endrizzi, M., Birren, B. & Lander, E. S. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature 423, 241–254 (2003)
Melcher, K. & Xu, H. E. Gal80–Gal80 interaction on adjacent Gal4p binding sites is required for complete GAL gene repression. EMBO J. 20, 841–851 (2001)
Wagner, A. Asymmetric functional divergence of duplicate genes in yeast. Mol. Biol. Evol. 19, 1760–1768 (2002)
Makova, K. D. & Li, W. H. Divergence in the spatial pattern of gene expression between human duplicate genes. Genome Res. 13, 1638–1645 (2003)
Gu, Z., Rifkin, S. A., White, K. P. & Li, W. H. Duplicate genes increase gene expression diversity within and between species. Nature Genet. 36, 577–579 (2004)
Huminiecki, L. & Wolfe, K. H. Divergence of spatial gene expression profiles following species-specific gene duplications in human and mouse. Genome Res. 14, 1870–1879 (2004)
Li, W. H., Yang, J. & Gu, X. Expression divergence between duplicate genes. Trends Genet. 21, 602–607 (2005)
Goodman, M., Moore, G. W. & Matsuda, G. Darwinian evolution in the genealogy of haemoglobin. Nature 253, 603–608 (1975)
Gould, A., Morrison, A., Sproat, G., White, R. A. & Krumlauf, R. Positive cross-regulation and enhancer sharing: two mechanisms for specifying overlapping Hox expression patterns. Genes Dev. 11, 900–913 (1997)
Cohen, B. A., Mitra, R. D., Hughes, J. D. & Church, G. M. A computational analysis of whole-genome expression data reveals chromosomal domains of gene expression. Nature Genet. 26, 183–186 (2000)
Hartl, D. L. & Clark, A. G. in Principles of Population Genetics 216 (Sinauer Associates, Sunderland, Massachusetts, 1997)
Brachmann, C. B. et al. Designer deletion strains derived from Saccharomyces cerevisiae S288C: a useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast 14, 115–132 (1998)
Kooistra, R., Hooykaas, P. J. & Steensma, H. Y. Efficient gene targeting in Kluyveromyces lactis . Yeast 21, 781–792 (2004)
Guldener, U., Heck, S., Fielder, T., Beinhauer, J. & Hegemann, J. H. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 24, 2519–2524 (1996)
Acknowledgements
We thank B. L. Williams for sharing unpublished data, reagents and the competition assay; J. E. Selegue for technical support; M. Johnston and Washington University in St Louis for access to laboratory space and equipment; Collection de Levures d'Intérêt Biotechnologique for K. lactis strain CLIB210; H. Y. Steensma for the NHEJ-deficient K. lactis strain and transformation protocol; J. H. Hegemann for the kanMX cassette; B. L. Williams, P. M. van Wynsberghe and Carroll laboratory members for technical advice; L. M. Olds for artwork; and B. L. Williams, B. Prud’homme and M. Johnston for critical reading of the manuscript. The Howard Hughes Medical Institute supported this work through an investigatorship (S.B.C.) and predoctoral fellowship (C.T.H.).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
The file contains Supplementary Discussion of Supplementary Tables S1-S4, Supplementary Tables S1-S4, Supplementary Figure S1 with Legend and additional references. (PDF 391 kb)
Rights and permissions
About this article
Cite this article
Hittinger, C., Carroll, S. Gene duplication and the adaptive evolution of a classic genetic switch. Nature 449, 677–681 (2007). https://doi.org/10.1038/nature06151
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature06151
This article is cited by
-
Explaining Human Diversity: the Need to Balance Fit and Complexity
Review of Philosophy and Psychology (2023)
-
From pairs of most similar sequences to phylogenetic best matches
Algorithms for Molecular Biology (2020)
-
Rapid functional divergence after small-scale gene duplication in grasses
BMC Evolutionary Biology (2019)
-
EMS1 and BRI1 control separate biological processes via extracellular domain diversity and intracellular domain conservation
Nature Communications (2019)
-
A naturally occurring variation in the BrMAM-3 gene is associated with aliphatic glucosinolate accumulation in Brassica rapa leaves
Horticulture Research (2018)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.