Abstract
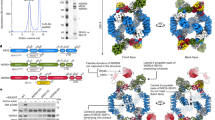
AMP-activated protein kinase (AMPK) is a central regulator of energy homeostasis in mammals and is an attractive target for drug discovery against diabetes, obesity and other diseases1,2,3,4,5. The AMPK homologue in Saccharomyces cerevisiae, known as SNF1, is essential for responses to glucose starvation as well as for other cellular processes, although SNF1 seems to be activated by a ligand other than AMP1,6,7,8. Here we report the crystal structure at 2.6 Å resolution of the heterotrimer core of SNF1. The ligand-binding site in the γ-subunit (Snf4) has clear structural differences from that of the Schizosaccharomyces pombe enzyme9, although our crystallographic data indicate that AMP can also bind to Snf4. The glycogen-binding domain in the β-subunit (Sip2) interacts with Snf4 in the heterotrimer but should still be able to bind carbohydrates10,11,12,13. Our structure is supported by a large body of biochemical and genetic data on this complex1,6,7,8,14,15,16,17,18. Most significantly, the structure reveals that part of the regulatory sequence in the α-subunit (Snf1)15,16,18,19 is sequestered by Snf4, demonstrating a direct interaction between the α- and γ-subunits and indicating that our structure may represent the heterotrimer core of SNF1 in its activated state.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout



Similar content being viewed by others
References
Kahn, B. B., Alquier, T., Carling, D. & Hardie, D. G. AMP-activated protein kinase: ancient energy gauge provides clues to modern understanding of metabolism. Cell Metab. 1, 15–25 (2005)
Hardie, D. G. & Sakamoto, K. AMPK: a key sensor of fuel and energy status in skeletal muscle. Physiology 21, 48–60 (2006)
Carling, D. AMP-activated protein kinase: balancing the scales. Biochimie 87, 87–91 (2005)
Kemp, B. E. et al. AMP-activated protein kinase, super metabolic regulator. Biochem. Soc. Trans. 31, 162–168 (2003)
Viollet, B. et al. Physiological role of AMP-activated protein kinase (AMPK): insights from knockout mouse models. Biochem. Soc. Trans. 31, 216–219 (2003)
Hardie, D. G., Carling, D. & Carlson, M. The AMP-activated/SNF1 protein kinase subfamily: metabolic sensors of the eukaryotic cell? Annu. Rev. Biochem. 67, 821–855 (1998)
Hong, S.-P. & Carlson, M. Regulation of Snf1 protein kinase in response to environmental stress. J. Biol. Chem. 282, 16838–16845 (2007)
Sanz, P. Snf1 protein kinase: a key player in the response to cellular stress in yeast. Biochem. Soc. Trans. 31, 178–181 (2003)
Townley, R. & Shapiro, L. Crystal structures of the adenylate sensor from fission yeast AMP-activated protein kinase. Science 315, 1726–1729 (2007)
Polekhina, G. et al. Structural basis for glycogen recognition by AMP-activated protein kinase. Structure 13, 1453–1462 (2005)
Polekhina, G. et al. AMPK β subunit targets metabolic stress sensing to glycogen. Curr. Biol. 13, 867–871 (2003)
Wiatrowski, H. A. et al. Mutations in the Gal83 glycogen-binding domain activate the Snf1/Gal83 kinase pathway by a glycogen-independent mechanism. Mol. Cell. Biol. 24, 352–361 (2004)
Hudson, E. R. et al. A novel domain in AMP-activated protein kinase causes glycogen storage bodies similar to those seen in hereditary cardiac arrhythmias. Curr. Biol. 13, 861–866 (2003)
Iseli, T. J. et al. AMP-activated protein kinase β subunit tethers α and γ subunits via its C-terminal sequence (186–270). J. Biol. Chem. 280, 13395–13400 (2005)
Jiang, R. & Carlson, M. Glucose regulates protein interactions within the yeast SNF1 protein kinase complex. Genes Dev. 10, 3105–3115 (1996)
Crute, B. E., Seefeld, K., Gamble, J., Kemp, B. E. & Witters, L. A. Functional domains of the α1 catalytic subunit of the AMP-activated protein kinase. J. Biol. Chem. 273, 35347–35354 (1998)
Jiang, R. & Carlson, M. The Snf1 protein kinase and its activating subunit, Snf4, interact with distinct domains of the Sip1/Sip2/Gal83 component in the kinase complex. Mol. Cell. Biol. 17, 2099–2106 (1997)
Leech, A., Nath, N., McCartney, R. R. & Schmidt, M. C. Isolation of mutations in the catalytic domain of the Snf1 kinase that render its activity independent of the Snf4 subunit. Eukaryot. Cell 2, 265–273 (2003)
Pang, T. et al. Conserved α-helix acts as autoinhibitory sequence in AMP-activated protein kinase α subunits. J. Biol. Chem. 282, 495–506 (2007)
Rudolph, M. J., Amodeo, G. A., Bai, Y. & Tong, L. Crystal structure of the protein kinase domain of yeast AMP-activated protein kinase Snf1. Biochem. Biophys. Res. Commun. 337, 1224–1228 (2005)
Nayak, V. et al. Structure and dimerization of the kinase domain from yeast Snf1, a member of the Snf1/AMPK protein family. Structure 14, 477–485 (2006)
Bateman, A. The structure of a domain common to archaebacteria and the homocystinuria disease protein. Trends Biochem. Sci. 22, 12–13 (1997)
Kemp, B. E. Bateman domains and adenosine derivatives form a binding contract. J. Clin. Invest. 113, 182–184 (2004)
Scott, J. W. et al. CBS domains form energy-sensing modules whose binding of adenosine ligands is disrupted by disease mutations. J. Clin. Invest. 113, 274–284 (2004)
Hamilton, S. R. et al. An activating mutation in the γ1 subunit of the AMP-activated protein kinase. FEBS Lett. 500, 163–168 (2001)
Rudolph, M. J. et al. Structure of the Bateman2 domain of yeast Snf4: dimeric association and relevance for AMP binding. Structure 15, 65–74 (2007)
Day, P. et al. Structure of a CBS-domain pair from the regulatory γ1 subunit of human AMPK in complex with AMP and ZMP. Acta Crystallogr. D 63, 587–596 (2007)
Adams, J. et al. Intrasteric control of AMPK via the γ1 subunit AMP allosteric regulatory site. Protein Sci. 13, 155–165 (2004)
Sanders, M. J., Grondin, P. O., Hegarty, B. D., Snowden, M. A. & Carling, D. Investigating the mechanism for AMP activation of the AMP-activated protein kinase cascade. Biochem. J. 403, 139–148 (2007)
Carson, M. Ribbon models of macromolecules. J. Mol. Graph. 5, 103–106 (1987)
Hendrickson, W. A., Horton, J. R. & LeMaster, D. M. Selenomethionyl proteins produced for analysis by multiwavelength anomalous diffraction (MAD): a vehicle for direct determination of three-dimensional structure. EMBO J. 9, 1665–1672 (1990)
Otwinowski, Z. & Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 (1997)
Storoni, L. C., McCoy, A. J. & Read, R. J. Likelihood-enhanced fast rotation functions. Acta Crystallogr. D 60, 432–438 (2004)
Townley, R. & Shapiro, L. Crystal structures of the adenylate sensor from fission yeast AMP-activated protein kinase. Science 315, 1726–1729 (2007)
Polekhina, G. et al. Structural basis for glycogen recognition by AMP-activated protein kinase. Structure 13, 1453–1462 (2005)
Brunger, A. T. et al. Crystallography & NMR System: A new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
Murshudov, G. N., Vagin, A. A. & Dodson, E. J. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D 53, 240–255 (1997)
Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47, 110–119 (1991)
Emsley, P. & Cowtan, K. D. Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004)
Acknowledgements
We thank M. Carlson for helpful discussions; H. Robinson and N. Whalen for setting up the X29A beamline; and J. Schwanof and R. Abramowitz for setting up the X4C beamline at the National Synchrotron Light Source. This research is supported in part by an NIH grant to L.T. G.A.A. was supported by an NIH training program in molecular biophysics.
The atomic coordinates are deposited at the Protein Data Bank under accession number 2QLV.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
Supplementary information
Supplementary Information
The file contains a more detailed Supplementary Methods section with additional references, Supplementary Table1 and Supplementary Figures 1-9 with Legends. (PDF 3690 kb)
Rights and permissions
About this article
Cite this article
Amodeo, G., Rudolph, M. & Tong, L. Crystal structure of the heterotrimer core of Saccharomyces cerevisiae AMPK homologue SNF1. Nature 449, 492–495 (2007). https://doi.org/10.1038/nature06127
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature06127
This article is cited by
-
RCO-3 and COL-26 form an external-to-internal module that regulates the dual-affinity glucose transport system in Neurospora crassa
Biotechnology for Biofuels (2021)
-
Functional analysis of CfSnf1 in the development and pathogenicity of anthracnose fungus Colletotrichum fructicola on tea-oil tree
BMC Genetics (2019)
-
Regulation and metabolic engineering strategies for permeases of Saccharomyces cerevisiae
World Journal of Microbiology and Biotechnology (2019)
-
Metabolomics profiling reveals differential adaptation of major energy metabolism pathways associated with autophagy upon oxygen and glucose reduction
Scientific Reports (2018)
-
Ketamine’s antidepressant effect is mediated by energy metabolism and antioxidant defense system
Scientific Reports (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.