Abstract
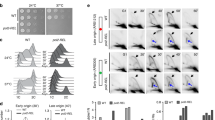
Specialized DNA polymerases (DNA pols) are required for lesion bypass in human cells1. Auxiliary factors have an important, but so far poorly understood, role. Here we analyse the effects of human proliferating cell nuclear antigen (PCNA) and replication protein A (RP-A) on six different human DNA pols—belonging to the B, Y and X classes—during in vitro bypass of different lesions. The mutagenic lesion 8-oxo-guanine (8-oxo-G) has high miscoding potential2,3,4. A major and specific effect was found for 8-oxo-G bypass with DNA pols λ and η. PCNA and RP-A allowed correct incorporation of dCTP opposite a 8-oxo-G template 1,200-fold more efficiently than the incorrect dATP by DNA pol λ, and 68-fold by DNA pol η, respectively. Experiments with DNA-pol-λ-null cell extracts suggested an important role for DNA pol λ. On the other hand, DNA pol ι, together with DNA pols α, δ and β, showed a much lower correct bypass efficiency. Our findings show the existence of an accurate mechanism to reduce the deleterious consequences of oxidative damage and, in addition, point to an important role for PCNA and RP-A in determining a functional hierarchy among different DNA pols in lesion bypass.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout


Similar content being viewed by others
References
Hubscher, U., Maga, G. & Spadari, S. Eukaryotic DNA polymerases. Annu. Rev. Biochem. 71, 133–163 (2002)
Collins, A. R. Oxidative DNA damage, antioxidants, and cancer. Bioessays 21, 238–246 (1999)
Krahn, J. M., Beard, W. A., Miller, H., Grollman, A. P. & Wilson, S. H. Structure of DNA polymerase β with the mutagenic DNA lesion 8-oxodeoxyguanine reveals structural insights into its coding potential. Structure 11, 121–127 (2003)
Shibutani, S., Takeshita, M. & Grollman, A. P. Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature 349, 431–434 (1991)
Zhang, Y. et al. Lesion bypass activities of human DNA polymerase μ. J. Biol. Chem. 277, 44582–44587 (2002)
Maga, G., Shevelev, I., Villani, G., Spadari, S. & Hubscher, U. Human replication protein A can suppress the intrinsic in vitro mutator phenotype of human DNA polymerase λ. Nucleic Acids Res. 34, 1405–1415 (2006)
Maga, G. et al. Human DNA polymerase λ functionally and physically interacts with proliferating cell nuclear antigen in normal and translesion DNA synthesis. J. Biol. Chem. 277, 48434–48440 (2002)
Maga, G. et al. DNA elongation by the human DNA polymerase λ polymerase and terminal transferase activities are differentially coordinated by proliferating cell nuclear antigen and replication protein A. J. Biol. Chem. 280, 1971–1981 (2005)
Haracska, L., Yu, S. L., Johnson, R. E., Prakash, L. & Prakash, S. Efficient and accurate replication in the presence of 7,8-dihydro-8-oxoguanine by DNA polymerase η. Nature Genet. 25, 458–461 (2000)
Vaisman, A. & Woodgate, R. Unique misinsertion specificity of pol ι may decrease the mutagenic potential of deaminated cytosines. EMBO J. 20, 6520–6529 (2001)
Maga, G., Frouin, I., Spadari, S. & Hubscher, U. Replication protein A as a “fidelity clamp” for DNA polymerase α. J. Biol. Chem. 276, 18235–18242 (2001)
Blanca, G. et al. Human DNA polymerases λ and β show different efficiencies of translesion DNA synthesis past abasic sites and alternative mechanisms for frameshift generation. Biochemistry 43, 11605–11615 (2004)
Haracska, L., Kondratick, C. M., Unk, I., Prakash, S. & Prakash, L. Interaction with PCNA is essential for yeast DNA polymerase η function. Mol. Cell 8, 407–415 (2001)
Haracska, L. et al. Targeting of human DNA polymerase ι to the replication machinery via interaction with PCNA. Proc. Natl Acad. Sci. USA 98, 14256–14261 (2001)
Hoffmann, J. S. et al. DNA polymerase β bypasses in vitro a single d(GpG)-cisplatin adduct placed on codon 13 of the HRAS gene. Proc. Natl Acad. Sci. USA 92, 5356–5360 (1995)
Bassett, E. et al. Frameshifts and deletions during in vitro translesion synthesis past Pt-DNA adducts by DNA polymerases β and η. DNA Repair (Amst.) 1, 1003–1016 (2002)
Weiser, T. et al. Biochemical and functional comparison of DNA polymerases α, δ and ε from calf thymus. J. Biol. Chem. 266, 10420–10428 (1991)
Bertocci, B., De Smet, A., Weill, J. C. & Reynaud, C. A. Nonoverlapping functions of DNA polymerases μ, λ, and terminal deoxynucleotidyltransferase during immunoglobulin V(D)J recombination in vivo. Immunity 25, 31–41 (2006)
Todaro, G. J. & Green, H. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J. Cell Biol. 17, 299–313 (1963)
Acknowledgements
We thank R. Woodgate for his generous gift of recombinant human DNA pol η and ι. U.H. and U.W. are supported by the Swiss National Science Foundation, by the UBS “im Auftrag eines Kunden”, and U.H. and E.F. by the University of Zürich, which gave a grant in aid to G.M. G.M. is supported partially by the CARIPLO Foundation Project “Oncogenetica e Proteomica della Replicazione”. G.V. is supported by CNRS and ARC.
Author Contributions G.M. and U.H. had the original idea. G.M. supervised the overall experimental strategy and performed all the experiments with human DNA pol λ and cell extracts; E.C. performed all the experiments with human DNA polymerases α, δ, β, ι and η; U.W. and B.B. generated the DNA POLL+/+ and POLL-/- MEFs and characterized their phenotype; G.V. provided the damaged templates; E.F. purified DNA pols α, δ, PCNA and RP-A; G.M., G.V. and U.H. designed and interpreted all the experiments and equally contributed to manuscript writing and figures preparation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests.
Supplementary information
Supplementary Information
This file contains Supplementary Figures S1-S5 with Legends and Supplementary Table S1. The Supplementary Figures provide additional evidences for the specificity of the observed effects of PCNA and RP-A on DNA polymerases lambda and eta during 8-oxo-G bypass, including results with other lesions such as abasic site and cis-platinum adduct.The Supplementary Table 1 summarizes all the kinetic constants for nucleotide incorporation by DNA polymerases lambda, alpha eta and iota opposite a normal G or an 8-oxo-G. (PDF 977 kb)
Rights and permissions
About this article
Cite this article
Maga, G., Villani, G., Crespan, E. et al. 8-oxo-guanine bypass by human DNA polymerases in the presence of auxiliary proteins. Nature 447, 606–608 (2007). https://doi.org/10.1038/nature05843
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature05843
This article is cited by
-
Distinctive roles of translesion polymerases DinB1 and DnaE2 in diversification of the mycobacterial genome through substitution and frameshift mutagenesis
Nature Communications (2022)
-
Analysis of DNA Polymerase λ Activity and Gene Expression in Response to Salt and Drought Stress in Oryza sativa Indica Rice Cultivars
Journal of Plant Growth Regulation (2022)
-
Structural basis of DNA synthesis opposite 8-oxoguanine by human PrimPol primase-polymerase
Nature Communications (2021)
-
Mutations induced by 8-hydroxyguanine (8-oxo-7,8-dihydroguanine), a representative oxidized base, in mammalian cells
Genes and Environment (2017)
-
Embryonic exposure to the widely-used herbicide atrazine disrupts meiosis and normal follicle formation in female mice
Scientific Reports (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.