Abstract
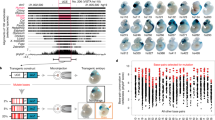
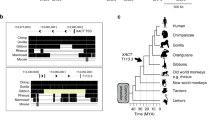
Hundreds of highly conserved distal cis-regulatory elements have been characterized so far in vertebrate genomes1. Many thousands more are predicted on the basis of comparative genomics2,3. However, in stark contrast to the genes that they regulate, in invertebrates virtually none of these regions can be traced by using sequence similarity, leaving their evolutionary origins obscure. Here we show that a class of conserved, primarily non-coding regions in tetrapods originated from a previously unknown short interspersed repetitive element (SINE) retroposon family that was active in the Sarcopterygii (lobe-finned fishes and terrestrial vertebrates) in the Silurian period at least 410 million years ago (ref. 4), and seems to be recently active in the ‘living fossil’ Indonesian coelacanth, Latimeria menadoensis. Using a mouse enhancer assay we show that one copy, 0.5 million bases from the neuro-developmental gene ISL1, is an enhancer that recapitulates multiple aspects of Isl1 expression patterns. Several other copies represent new, possibly regulatory, alternatively spliced exons in the middle of pre-existing Sarcopterygian genes. One of these, a more than 200-base-pair ultraconserved region5, 100% identical in mammals, and 80% identical to the coelacanth SINE, contains a 31-amino-acid-residue alternatively spliced exon of the messenger RNA processing gene PCBP2 (ref. 6). These add to a growing list of examples7 in which relics of transposable elements have acquired a function that serves their host, a process termed ‘exaptation’8, and provide an origin for at least some of the many highly conserved vertebrate-specific genomic sequences.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Woolfe, A. et al. Highly conserved non-coding sequences are associated with vertebrate development. PLoS Biol. 3, e7 (2005)
Siepel, A. et al. Evolutionarily conserved elements in vertebrate, insect, worm, and yeast genomes. Genome Res. 15, 1034–1050 (2005)
International Mouse Genome Sequencing Consortium. Initial sequencing and comparative analysis of the mouse genome. Nature 420, 520–562 (2002)
Graur, D. & Li, W.-H. Fundamentals of Molecular Evolution 2nd edn, Appendix I (Sinauer, Sunderland, Massachusetts, 2000)
Bejerano, G. et al. Ultraconserved elements in the human genome. Science 304, 1321–1325 (2004)
Makeyev, A. V. & Liebhaber, S. A. The poly(C)-binding proteins: A multiplicity of functions and a search for mechanisms. RNA 8, 265–278 (2002)
Brosius, J. The contribution of RNAs and retroposition to evolutionary novelties. Genetica 118, 99–116 (2003)
Gould, S. & Vrba, E. Exaptation—a missing term in the science of form. Paleobiology 8, 4–15 (1982)
Bejerano, G., Haussler, D. & Blanchette, M. Into the heart of darkness: Large-scale clustering of human non-coding DNA. Bioinformatics 20, i40–i48 (2004)
Weiner, A. M. SINEs and LINEs: The art of biting the hand that feeds you. Curr. Opin. Cell Biol. 14, 343–350 (2002)
Deininger, P. L. & Batzer, M. A. Mammalian retroelements. Genome Res. 12, 1455–1465 (2002)
Pfaff, S. L., Mendelsohn, M., Stewart, C. L., Edlund, T. & Jessell, T. M. Requirement for LIM homeobox gene Isl1 in motor neuron generation reveals a motor neuron-dependent step in interneuron differentiation. Cell 84, 309–320 (1996)
Uemura, O. et al. Comparative functional genomics revealed conservation and diversification of three enhancers of the isl1 gene for motor and sensory neuron-specific expression. Dev. Biol. 278, 587–606 (2005)
Caton, A. et al. The branchial arches and HGF are growth-promoting and chemoattractant for cranial motor axons. Development 127, 1751–1766 (2000)
Lev-Maor, G., Sorek, R., Shomron, N. & Ast, G. The birth of an alternatively spliced exon: 3′ splice-site selection in Alu exons. Science 300, 1288–1291 (2003)
Makalowski, W. Genomics. Not junk after all. Science 300, 1246–1247 (2003)
Lewis, B. P., Green, R. E. & Brenner, S. E. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc. Natl Acad. Sci. USA 100, 189–192 (2003)
Chkheidze, A. N. & Liebhaber, S. A. A novel set of nuclear localization signals determine distributions of the alphaCP RNA-binding proteins. Mol. Cell. Biol. 23, 8405–8415 (2003)
Kim, J. H., Hahm, B., Kim, Y. K., Choi, M. & Jang, S. K. Protein–protein interaction among hnRNPs shuttling between nucleus and cytoplasm. J. Mol. Biol. 298, 395–405 (2000)
Waggoner, S. A. & Liebhaber, S. A. Identification of mRNAs associated with αCP2-containing RNP complexes. Mol. Cell. Biol. 23, 7055–7067 (2003)
Gunduz, E. et al. Genetic and epigenetic alterations of BRG1 promote oral cancer development. Int. J. Oncol. 26, 201–210 (2005)
Li, Y., Lu, W. & Bu, G. Striking differences of LDL receptor-related protein 1B expression in mouse and human. Biochem. Biophys. Res. Commun. 333, 868–873 (2005)
McClintock, B. The origin and behavior of mutable loci in maize. Proc. Natl Acad. Sci. USA 36, 344–355 (1950)
Britten, R. J. & Davidson, E. H. Repetitive and non-repetitive DNA sequences and a speculation on the origins of evolutionary novelty. Q. Rev. Biol. 46, 111–138 (1971)
Kazazian, H. H. Jr . Mobile elements: Drivers of genome evolution. Science 303, 1626–1632 (2004)
McEwen, G. K. et al. Ancient duplicated conserved noncoding elements in vertebrates: A genomic and functional analysis. Genome Res. doi:10.1101/gr.4143406 (2006)
Danke, J. et al. Genome resource for the Indonesian coelacanth, Latimeria menadoensis. J. Exp. Zoolog. A 301, 228–234 (2004)
Kothary, R. et al. A transgene containing lacZ inserted into the dystonia locus is expressed in neural tube. Nature 335, 435–437 (1988)
Poulin, F. et al. In vivo characterization of a vertebrate ultraconserved enhancer. Genomics 85, 774–781 (2005)
Feldheim, D. A. et al. Topographic guidance labels in a sensory projection to the forebrain. Neuron 21, 1303–1313 (1998)
Acknowledgements
We thank the various sequencing consortia and research groups for the numerous genomic regions used in this study; A. Hinrichs, M. Diekhans, R. Harte, G. Barber and the UCSC browser team, M. Shoukry, I. Plajzer-Frick, S. Chanan and V. Afzal for technical help; R. Baertsch, T. Furey, E. Margulies and J. S. Pedersen for sharing unpublished data; and W. Miller, M. Blanchette, D. Feldheim, M. Nobrega, M. Ares, C. Sugnet, M. Dermitzakis and J. Brosius for discussions. Research conducted at University of California Santa Cruz was supported by the National Human Genome Research Institute and the Howard Hughes Medical Institute; Research conducted at the E.O. Lawrence Berkeley National Laboratory was supported by grants from the Programs for Genomic Application, the NHLBI and performed under a Department of Energy Contract, University of California.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests.
Supplementary information
Supplementary Notes
This file contains Supplementary Methods, Supplementary Discussion, Supplementary Figures 1–15 and Supplementary Tables 1–9. (PDF 1641 kb)
Rights and permissions
About this article
Cite this article
Bejerano, G., Lowe, C., Ahituv, N. et al. A distal enhancer and an ultraconserved exon are derived from a novel retroposon. Nature 441, 87–90 (2006). https://doi.org/10.1038/nature04696
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature04696
This article is cited by
-
Transposable elements as essential elements in the control of gene expression
Mobile DNA (2023)
-
The genome of the glasshouse plant noble rhubarb (Rheum nobile) provides a window into alpine adaptation
Communications Biology (2023)
-
Transposable elements in mammalian chromatin organization
Nature Reviews Genetics (2023)
-
Origin, evolution, and tissue-specific functions of the porcine repetitive element 1
Genetics Selection Evolution (2022)
-
Remnants of SIRE1 retrotransposons in human genome?
Journal of Genetics (2022)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.