Abstract
Arising from: C. P. Groves, G. B. Schaller, G. Amato & K. Khounboline Nature 386, 335 (1997)
In 1997, the rediscovery of Sus bucculentus in Laos was announced by Groves et al.1 — this wild pig species had gone unrecorded since first being described in 1892. Although the identification of the new specimen was based initially on morphology, the authors also used a 7% sequence divergence from the common Eurasian pig S. scrofa (based on their analysis of 327 base pairs of the gene encoding mitochondrial 12S ribosomal RNA) as support for the species status of S. bucculentus. Concerned about the large divergence reported for a relatively conserved gene, and the absence of the sequence in any public database, we analysed an additional tissue sample from the specimen and found only 0.6% divergence from S. scrofa. Our more extensive analysis places the sample within the S. scrofa clade, calling into question the species status of S. bucculentus and demonstrating the need for both phylogenetic and morphological evidence in defining species.
Similar content being viewed by others
Main
We extracted DNA from a dried tissue sample of the more recently described S. bucculentus specimen1 (kindly supplied by C. P. Groves). The D-loop, 12S and cytochrome b regions of mitochondrial DNA were amplified, sequenced and compared with a wide range of sequences from GenBank and from our laboratory database. The comparison included three samples of S. scrofa from archaeological sites on Pacific islands; these were extracted in a dedicated ancient-DNA laboratory using standard precautions to avoid contamination2, and were independently replicated by J. Hay (Massey University, New Zealand) in an ancient-DNA laboratory in which porcine DNA had not previously been processed.
Phylogenetic analysis of the 12S-rRNA gene would be uninformative because of poor species representation in GenBank, so we used a barcode approach3 to compare intraspecific and interspecific sequence distances4. For cytochrome b and D-loop, phylogenetic trees were estimated using both bayesian5 and maximum-likelihood6 methods.
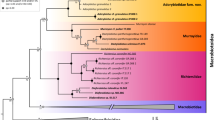
For the 12S-rRNA gene, the mean pairwise distance between S. bucculentus and S. scrofa (0.006) is identical to that within S. scrofa (0.006), and only a tenth of the pairwise distance (0.055) between S. scrofa and the African warthog (Phacochoerus africanus). Unlike the 7% sequence difference reported earlier1, this 0.6% difference is consistent with the conserved nature of the 12S-rRNA gene. For cytochrome b and D-loop, all four phylogenetic trees have similar topology, placing S. bucculentus within the Asian subclade of S. scrofa. Furthermore, in the D-loop analyses (Fig. 1), S. bucculentus sits in a clade that includes S. scrofa from Thailand and Myanmar, countries that border Laos. Coincidentally, an archaeological specimen from Tikopia in the Solomon Islands, albeit from an uncertain chronological context7, had a D-loop sequence identical to that of S. bucculentus.
Triangles represent monophyletic clades and circles represent paraphyletic groups and single sequences. Clades are coloured and named according to ref. 9. Nodes marked by asterisks have posterior P < 80%. All clades represent S. scrofa except when labelled: b, S. barbatus; ce, S. celebensis; cr, S. cristatus; s, S. scrofa; v, S. verrucosus. GenBank accession numbers for the new sequences used in this study are DQ444703–DQ444711. Sample details, methods of DNA extraction, amplification and sequencing, computational methods, the full phylogenetic trees based on the D-loop and cytochrome b regions of the mitochondrial DNA, and the barcoding analysis of the 12S gene are available from the authors (http://www.cebl.auckland.ac.nz/~hros001/Susbucc/).
These results undermine the species status of S. bucculentus. The partial skull from 1997 was morphologically identical to the 1892 skull1. But Groves and colleagues have recently judged the key taxonomic characters of the mandibular canine and malar tuberosity to be less distinctive than once thought, even though multivariate analyses of cranial and dental measurements tend to separate the two skulls from samples of Indochinese S. scrofa8. Our phylogenetic results are consistent with a recently shared ancestry between the 1997 S. bucculentus sample from the Annamite Range and Asian domestic and wild S. scrofa. Also, our S. bucculentus 12S-rRNA gene sequence was within the range of intraspecific variation shown by S. scrofa.
There are three possible explanations for these findings, a view shared by C. P. Groves (personal communication). First, hybridization may have introduced a locally occurring mitochondrial haplotype from S. scrofa into S. bucculentus. Second, speciation and morphological differentiation of S. bucculentus may have occurred relatively recently and so subsequent mitochondrial lineage sorting remains incomplete. A third possibility, however, is that the morphology previously1,8 designated as that of a separate species, S. bucculentus, is no more than one extreme of the natural range of variation shown by the wild Indochinese S. scrofa.
References
Groves, C. P., Schaller, G. B., Amato, G. & Khounboline, K. Nature 386, 335 (1997).
Handt, O., Höss, M., Krings, M. & Pääbo, S. Experientia 50, 524–529 (1994).
Hebert, P. D. N., Cywinska, A., Ball, S. L. & deWaard, J. R. Proc. R. Soc. Lond. B 270, 313–321 (2003).
Swofford, D. L. PAUP*: Phylogenetic Analysis Using Parsimony (* and Other Methods) (Sinauer, Sunderland, Massachusetts, 2002).
Huelsenbeck, J. P. & Ronquist, F. Bioinformatics 17, 754–755 (2001).
Guindon, S. & Gascuel, O. Syst. Biol. 52, 696–704 (2003).
Kirch, P. V. & Yen, D. E. in Tikopia: The Prehistory and Ecology of a Polynesian Outlier 100 (Bishop Museum, Honolulu, 1982).
Groves, C. P. & Schaller, G. B. in Antelopes, Deer, and Relatives (eds Vrba, E. S. & Schaller, G. B.) 261–282 (Yale Univ. Press, New Haven and London, 2000).
Larson, G. et al. Science 307, 1618–1621 (2005).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Robins, J., Ross, H., Allen, M. et al. Sus bucculentus revisited. Nature 440, E7 (2006). https://doi.org/10.1038/nature04770
Published:
Issue Date:
DOI: https://doi.org/10.1038/nature04770
This article is cited by
-
A Sealers Midden Provides Evidence a Live Pig (Sus scrofa) was Taken Ashore at Heard Island During the “Elephanting” Industry (1855–1882)
Journal of Maritime Archaeology (2012)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.