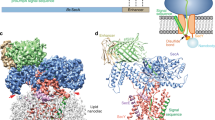
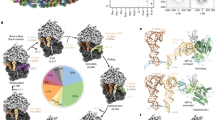
Abstract
Secreted and membrane proteins are translocated across or into cell membranes through a protein-conducting channel (PCC). Here we present a cryo-electron microscopy reconstruction of the Escherichia coli PCC, SecYEG, complexed with the ribosome and a nascent chain containing a signal anchor. This reconstruction shows a messenger RNA, three transfer RNAs, the nascent chain, and detailed features of both a translocating PCC and a second, non-translocating PCC bound to mRNA hairpins. The translocating PCC forms connections with ribosomal RNA hairpins on two sides and ribosomal proteins at the back, leaving a frontal opening. Normal mode-based flexible fitting of the archaeal SecYEβ structure into the PCC electron microscopy densities favours a front-to-front arrangement of two SecYEG complexes in the PCC, and supports channel formation by the opening of two linked SecY halves during polypeptide translocation. On the basis of our observation in the translocating PCC of two segregated pores with different degrees of access to bulk lipid, we propose a model for co-translational protein translocation.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Simon, S. M. & Blobel, G. A protein-conducting channel in the endoplasmic reticulum. Cell 65, 371–380 (1991)
Wickner, W., Driessen, A. J. M. & Hartl, F. U. The enzymology of protein translocation across the Escherichia coli plasma membrane. Annu. Rev. Biochem. 60, 101–124 (1991)
Brundage, L. et al. The purified E. coli integral membrane protein SecY/E is sufficient for reconstitution of SecA-dependent precursor protein translocation. Cell 62, 649–657 (1990)
Gorlich, D. & Rapoport, T. A. Protein translocation into proteoliposomes reconstituted from purified components of the endoplasmic reticulum membrane. Cell 75, 615–630 (1993)
Gilmore, R. & Blobel, G. Translocation of secretory proteins across the microsomal membrane occurs through an environment accessible to aqueous perturbants. Cell 42, 497–505 (1985)
Simon, S. M., Blobel, G. & Zimmerberg, J. Large aqueous channels in membrane vesicles derived from the rough endoplasmic reticulum of canine pancreas or the plasma membrane of Escherichia coli. Proc. Natl Acad. Sci. USA 86, 6176–6180 (1989)
Mothes, W. et al. Molecular mechanism of membrane protein integration into the endoplasmic reticulum. Cell 89, 523–533 (1997)
Hessa, T. et al. Recognition of transmembrane helices by the endoplasmic reticulum translocon. Nature 433, 377–381 (2005)
Beckmann, R. et al. Alignment of conduits for the nascent polypeptide chain in the ribosome–Sec61 complex. Science 278, 2123–2126 (1997)
Menetret, J.-F. et al. The structure of ribosome–channel complexes engaged in protein translocation. Mol. Cell 6, 1219–1232 (2000)
Beckmann, R. et al. Architecture of the protein-conducting channel associated with the translating 80S ribosome. Cell 107, 361–372 (2001)
Morgan, D. G. et al. Structure of the mammalian ribosome-channel complex at 17 Å resolution. J. Mol. Biol. 324, 871–886 (2002)
van den Berg, B. et al. X-ray structure of a protein-conducting channel. Nature 427, 36–44 (2004)
Rapoport, T. A., Goder, V., Heinrich, S. U. & Matlack, K. E. Membrane-protein integration and the role of the translocation channel. Trends Cell Biol. 14, 568–575 (2004)
Prinz, A. et al. Evolutionarily conserved binding of ribosomes to the translocation channel via the large ribosomal tRNA. EMBO J. 19, 1900–1906 (2000)
Raden, D., Song, W. & Gilmore, R. Role of the cytoplasmic segments of Sec61α in the ribosome-binding and translocation-promoting activities of the Sec61 complex. J. Cell Biol. 150, 53–64 (2000)
Cheng, Z., Jiang, Y., Mandon, E. C. & Gilmore, R. Identification of cytoplasmic residues of Sec61p involved in ribosome binding and cotranslational translocation. J. Cell Biol. 168, 67–77 (2005)
Nakatogawa, H. & Ito, K. The ribosomal exit tunnel functions as a discriminating gate. Cell 108, 629–636 (2002)
Bessonneau, P., Besson, V., Collinson, I. & Duong, F. The SecYEG preprotein translocation channel is a conformationally dynamic and dimeric structure. EMBO J. 21, 995–1003 (2002)
van der Sluis, E. O., Nouwen, N. & Driessen, A. J. M. SecY–SecY and SecY–SecG contacts revealed by site-specific crosslinking. FEBS Lett. 527, 159–165 (2002)
Tama, F., Miyashita, O. & Brooks, C. L. III NMFF: Flexible high-resolution annotation of low-resolution experimental data from cryo-EM maps using normal mode analysis. J. Struct. Biol. 147, 315–326 (2004)
Go, N., Noguti, T. & Nishikawa, T. Dynamics of a small globular protein in terms of low-frequency vibrational modes. Proc. Natl Acad. Sci. USA 80, 3696–3700 (1983)
Kaufmann, A. et al. Cysteine-directed cross-linking demonstrates that helix 3 of SecE is close to helix 2 of SecY and helix 3 of a neighbouring SecE. Biochemistry 38, 9115–9125 (1999)
Veenendaal, A., van der Does, C. & Driessen, A. Mapping the sites of interaction between SecY and SecE by cysteine scanning mutagenesis. J. Biol. Chem. 276, 32559–32566 (2001)
Tani, K., Tokuda, H. & Mizushima, S. Translocation of ProOmpA possessing an intramolecular disulfide bridge into membrane vesicles of Escherichia coli. Effect of membrane energization. J. Biol. Chem. 265, 17341–17347 (1990)
Wirth, A. et al. The Sec61p complex is a dynamic precursor activated channel. Mol. Cell 12, 261–268 (2003)
Martoglio, B., Hofmann, M. W., Brunner, J. & Dobberstein, B. The protein-conducting channel in the membrane of the endoplasmic reticulum is open laterally toward the lipid bilayer. Cell 81, 207–214 (1995)
Levy, R., Wiedmann, M. & Kreibich, G. In vitro binding of ribosomes to the β subunit of the Sec61p protein translocation complex. J. Biol. Chem. 276, 2340–2346 (2001)
Nishiyama, K.-i., Mizushima, S. & Tokuda, H. A novel membrane protein involved in protein translocation across the cytoplasmic membrane of Escherichia coli. EMBO J. 12, 3409–3415 (1993)
Schatz, P. J. et al. One of three transmembrane stretches is sufficient for the functioning of the SecE protein, a membrane component of the E. coli secretion machinery. EMBO J. 10, 1749–1757 (1991)
Kalies, K. U., Rapoport, T. A. & Hartmann, E. The β subunit of the Sec61 complex facilitates cotranslational protein transport and interacts with the signal peptidase during translocation. J. Cell Biol. 141, 887–894 (1998)
Plath, K. et al. Signal sequence recognition in posttranslational protein transport across the yeast ER membrane. Cell 94, 795–807 (1998)
Laird, V. & High, S. Discrete cross-linking products identified during membrane protein biosynthesis. J. Biol. Chem. 272, 1983–1989 (1997)
Scotti, P. A. et al. YidC, the E. coli homologue of mitochondrial Oxa1p, is a component of the Sec translocase. EMBO J. 19, 542–549 (2000)
High, S. et al. Site-specific photocross-linking reveals that Sec61p and TRAM contact different regions of a membrane-inserted signal sequence. J. Biol. Chem. 268, 26745–26751 (1993)
Valent, Q. A. et al. The Escherichia coli SRP and SecB targeting pathways converge at the translocon. EMBO J. 17, 2504–2512 (1998)
Neumann-Haefelin, C., Schafer, U., Muller, M. & Koch, H. G. SRP-dependent cotranslational targeting and SecA-dependent translocation analyzed as individual steps in the export of a bacterial protein. EMBO J. 19, 6419–6426 (2000)
Zito, C. R. & Oliver, D. Two-stage binding of SecA to the bacterial translocon regulates ribosome–translocon interaction. J. Biol. Chem. 278, 40640–40646 (2003)
Wagenknecht, T., Grassucci, R. & Frank, J. Electron microscopy and computer image averaging of ice-embedded large ribosomal subunits from Escherichia coli. J. Mol. Biol. 199, 137–147 (1988)
Frank, J. et al. SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J. Struct. Biol. 116, 190–199 (1996)
Valle, M. et al. Cryo-EM reveals an active role for aminoacyl-tRNA in the accommodation process. EMBO J. 21, 3557–3567 (2002)
Gabashvili, I. S. et al. Solution structure of the E. coli 70S ribosome at 11.5 Å resolution. Cell 100, 537–549 (2000)
Chapman, M. S. Restrained real-space macromolecular atomic refinement using a new resolution-dependent electron density function. Acta Crystallogr. A 51, 69–80 (1995)
Tronrud, D. E., Ten Eyck, L. F. & Matthews, B. W. An efficient general-purpose least-squares refinement program for macromolecular structures. Acta Crystallogr. A 43, 489–501 (1987)
Gao, H. et al. Study of the structural dynamics of the E. coli 70S ribosome using real-space refinement. Cell 113, 789–801 (2003)
Harms, J. M. et al. Alterations at the peptidyl transferase centre of the ribosome induced by the synergistic action of the streptogramins dalfopristin and quinupristin. BMC Biol. 2, 4 (2004)
Tirion, M. M. Large amplitude elastic motions in proteins from a single-parameter, atomic analysis. Phys. Rev. Lett. 77, 1905–1908 (1996)
Breyton, C. et al. Three-dimensional structure of the bacterial protein-translocation complex SecYEG. Nature 418, 662–665 (2002)
Brunger, A. T. et al. Crystallography NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D 54, 905–921 (1998)
Acknowledgements
We thank R. A. Grassucci for training on the Tecnai F30 electron microscope; G. S. Allen for assistance with supervised classification, the software package O, and discussion of the manuscript; and M. Watters for assistance with the illustrations. N.B. thanks I. Berger and T. Ishikawa for advice on sample preparation and cryo-EM density symmetry analysis. This work was supported by Howard Hughes Medical Institute, National Science Foundation and National Institutes of Health (NIH) grants to J.F., and a Multiscale Modeling Tools for Structural Biology grant, funded by the NIH, to C.L.B. III. N.B. was supported by a grant from the Swiss National Science Foundation (SNSF), the NCCR Structural Biology program of the SNSF and a Young Investigator grant from the Human Frontier Science Program. C.S. was supported by post-doctoral fellowships from the Roche Research Foundation and the Ernst Schering Research Foundation. Author Contributions Grid preparation, cryo-EM, data processing, and atomic model generation, fitting, refinement and interpretation were done by K.M. (in the laboratory of J.F.). E. coli SecYEG–RNC complex preparation and PCC electron microscopy density symmetry analysis was performed by C.S. and S.J. (in the laboratory of N.B.). T.S. (J.F.) assisted in cryo-EM data processing. F.T. (in the laboratory of C.L.B. III) performed the NMA and NMFF.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
Coordinates for the translocating and non-translocating PCC have been deposited in the RCSB Protein Data Bank, with the accession codes 2AKI and 2AKH, respectively. The cryo-EM map of the E. coli RNC–SecYEG complex has been deposited in the EBI Macromolecular Structure Database, with the accession code EMD-1143. Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests.
Supplementary information
Supplementary Figure 1
Generation of RNC-PCC complexes. (PDF 68 kb)
Supplementary Figure 2
Selfrotation analysis of the non-translocating PCC EM density using the program GLRF. (PDF 268 kb)
Supplementary Figure 3
Normal mode analysis of the plug-less SecY complex. (PDF 1224 kb)
Supplementary Figure 4
Normal mode-based flexible fitting (NMFF) of the plug-less SecY complex. (PDF 1046 kb)
Supplementary Figure 5
Effect of resolution on the cryo-EM density of the translocating PCC. (PDF 73 kb)
Supplementary Notes
This file contains the Supplementary Results, Supplementary Discussion, Supplementary Figure Legends, Supplementary Methods and additional references. (DOC 76 kb)
Rights and permissions
About this article
Cite this article
Mitra, K., Schaffitzel, C., Shaikh, T. et al. Structure of the E. coli protein-conducting channel bound to a translating ribosome. Nature 438, 318–324 (2005). https://doi.org/10.1038/nature04133
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature04133
This article is cited by
-
Strategies to enhance soluble production of heterologous proteins in Escherichia coli
Biologia (2022)
-
Protein Transport Across the Bacterial Plasma Membrane by the Sec Pathway
The Protein Journal (2019)
-
Protein export through the bacterial Sec pathway
Nature Reviews Microbiology (2017)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.