Abstract
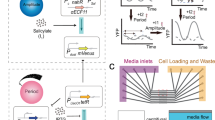
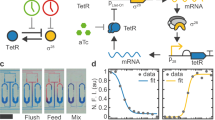
Autonomous oscillations found in gene expression and metabolic, cardiac and neuronal systems1,2,3,4 have attracted significant attention both because of their obvious biological roles and their intriguing dynamics. In addition, de novo designed5,6,7,8,9,10,11,12 oscillators13,14 have been demonstrated, using components that are not part of the natural oscillators. Such oscillators are useful in testing the design principles and in exploring potential applications not limited by natural cellular behaviour15. To achieve transcriptional and metabolic integration characteristic of natural oscillators, here we designed and constructed a synthetic circuit in Escherichia coli K12, using glycolytic flux to generate oscillation through the signalling metabolite acetyl phosphate. If two metabolite pools are interconverted by two enzymes that are placed under the transcriptional control of acetyl phosphate, the system oscillates when the glycolytic rate exceeds a critical value. We used bifurcation analysis to identify the boundaries of oscillation, and verified these experimentally. This work demonstrates the possibility of using metabolic flux as a control factor in system-wide oscillation, as well as the predictability of a de novo gene–metabolic circuit designed using nonlinear dynamic analysis.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Goldbeter, A. Biochemical Oscillations and Cellular Rhythms: The Molecular Bases of Periodic and Chaotic Behaviour (Cambridge Univ. Press, New York, 1996)
Hess, B. Periodic patterns in biology. Naturwissenschaften 87, 199–211 (2000)
Bier, M., Teusink, B., Kholodenko, B. N. & Westerhoff, H. V. Control analysis of glycolytic oscillations. Biophys. Chem. 62, 15–24 (1996)
Bier, M., Bakker, B. M. & Westerhoff, H. V. How yeast cells synchronize their glycolytic oscillations: a perturbation analytical treatment. Biophys. J. 78, 1087–1093 (2000)
Farmer, W. R. & Liao, J. C. Improving lycopene production in Escherichia coli by engineering metabolic control. Nature Biotechnol. 18, 533–537 (2000)
Gardner, T. S., Cantor, C. R. & Collins, J. J. Construction of a genetic toggle switch in Escherichia coli. Nature 403, 339–342 (2000)
Becskei, A., Seraphin, B. & Serrano, L. Positive feedback in eukaryotic gene networks: cell differentiation by graded to binary response conversion. EMBO J. 20, 2528–2535 (2001)
Guet, C. C., Elowitz, M. B., Hsing, W. & Leibler, S. Combinatorial synthesis of genetic networks. Science 296, 1466–1470 (2002)
Isaacs, F. J., Hasty, J., Cantor, C. R. & Collins, J. J. Prediction and measurement of an autoregulatory genetic module. Proc. Natl Acad. Sci. USA 100, 7714–7719 (2003)
Basu, S., Mehreja, R., Thiberge, S., Chen, M. T. & Weiss, R. Spatiotemporal control of gene expression with pulse-generating networks. Proc. Natl Acad. Sci. USA 101, 6355–6360 (2004)
Bulter, T. et al. Design of artificial cell–cell communication using gene and metabolic networks. Proc. Natl Acad. Sci. USA 101, 2299–2304 (2004)
You, L., Cox, R. S. III, Weiss, R. & Arnold, F. H. Programmed population control by cell–cell communication and regulated killing. Nature 428, 868–871 (2004)
Elowitz, M. B. & Leibler, S. A synthetic oscillatory network of transcriptional regulators. Nature 403, 335–338 (2000)
Atkinson, M. R., Savageau, M. A., Myers, J. T. & Ninfa, A. J. Development of genetic circuitry exhibiting toggle switch or oscillatory behavior in Escherichia coli. Cell 113, 597–607 (2003)
Kobayashi, H. et al. Programmable cells: interfacing natural and engineered gene networks. Proc. Natl Acad. Sci. USA 101, 8414–8419 (2004)
Kumari, S. et al. Regulation of acetyl coenzyme A synthetase in Escherichia coli. J. Bacteriol. 182, 4173–4179 (2000)
McCleary, W. R. & Stock, J. B. Acetyl phosphate and the activation of two-component response regulators. J. Biol. Chem. 269, 31567–31572 (1994)
Lutz, R. & Bujard, H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1–I2 regulatory elements. Nucleic Acids Res. 25, 1203–1210 (1997)
Andersen, J. B. et al. New unstable variants of green fluorescent protein for studies of transient gene expression in bacteria. Appl. Environ. Microbiol. 64, 2240–2246 (1998)
Feng, J. et al. Role of phosphorylated metabolic intermediates in the regulation of glutamine synthetase synthesis in Escherichia coli. J. Bacteriol. 174, 6061–6070 (1992)
Haldimann, A. & Wanner, B. L. Conditional-replication, integration, excision, and retrieval plasmid-host systems for gene structure-function studies of bacteria. J. Bacteriol. 183, 6384–6393 (2001)
Rutter, J., Reick, M. & McKnight, S. L. Metabolism and the control of circadian rhythms. Annu. Rev. Biochem. 71, 307–331 (2002)
Rutter, J., Reick, M., Wu, L. C. & McKnight, S. L. Regulation of clock and NPAS2 DNA binding by the redox state of NAD cofactors. Science 293, 510–514 (2001)
Kaasik, K. & Lee, C. C. Reciprocal regulation of haem biosynthesis and the circadian clock in mammals. Nature 430, 467–471 (2004)
Ferry, G. et al. Substrate specificity and inhibition studies of human serotonin N-acetyltransferase. J. Biol. Chem. 275, 8794–8805 (2000)
Brosius, J. & Holy, A. Regulation of ribosomal RNA promoters with a synthetic lac operator. Proc. Natl Acad. Sci. USA 81, 6929–6933 (1984)
Keiler, K. C., Waller, P. R. & Sauer, R. T. Role of a peptide tagging system in degradation of proteins synthesized from damaged messenger RNA. Science 271, 990–993 (1996)
Rines, D. R., He, X. & Sorger, P. K. Quantitative microscopy of green fluorescent protein-labeled yeast. Methods Enzymol. 351, 16–34 (2002)
Iooss, G. & Joseph, D. D. Elementary Stability and Bifurcation Theory (Springer-Verlag, New York, 1989)
Acknowledgements
The authors thank V. Roychowdhury and J. Bridgewater for discussions. This work was partially funded by the Center for Cell Mimetic Space Exploration, a National Aeronautics and Space Administration University Research, Engineering, and Technology Institute. J.C.L. is a member of California NanoSystems Institute and UCLA-DOE Institute for Genomics and Proteomics.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Supplementary information
Supplementary Notes
Contains Supplementary Methods and Supplementary Equations. Also includes Supplementary Table S1, Supplementary Figure S1 and additional references. (PDF 261 kb)
Rights and permissions
About this article
Cite this article
Fung, E., Wong, W., Suen, J. et al. A synthetic gene–metabolic oscillator. Nature 435, 118–122 (2005). https://doi.org/10.1038/nature03508
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature03508
This article is cited by
-
Using a synthetic machinery to improve carbon yield with acetylphosphate as the core
Nature Communications (2023)
-
Integration of photocatalytic and dark-operating catalytic biomimetic transformations through DNA-based constitutional dynamic networks
Nature Communications (2021)
-
Controlling biocatalytic cascades with enzyme–DNA dynamic networks
Nature Catalysis (2020)
-
Predictive biology: modelling, understanding and harnessing microbial complexity
Nature Reviews Microbiology (2020)
-
Massive computational acceleration by using neural networks to emulate mechanism-based biological models
Nature Communications (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.