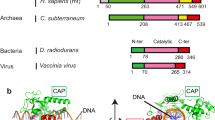
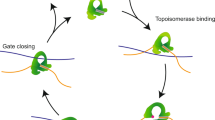
Abstract
Topoisomerases relieve the torsional strain in DNA that is built up during replication and transcription. They are vital for cell proliferation1,2,3 and are a target for poisoning by anti-cancer drugs4,5. Type IB topoisomerase (TopIB) forms a protein clamp around the DNA duplex6,7,8 and creates a transient nick that permits removal of supercoils. Using real-time single-molecule observation, we show that TopIB releases supercoils by a swivel mechanism that involves friction between the rotating DNA and the enzyme cavity: that is, the DNA does not freely rotate. Unlike a nicking enzyme, TopIB does not release all the supercoils at once, but it typically does so in multiple steps. The number of supercoils removed per step follows an exponential distribution. The enzyme is found to be torque-sensitive, as the mean number of supercoils per step increases with the torque stored in the DNA. We propose a model for topoisomerization in which the torque drives the DNA rotation over a rugged periodic energy landscape in which the topoisomerase has a small but quantifiable probability to religate the DNA once per turn.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Champoux, J. J. DNA topoisomerases: structure, function, and mechanism. Annu. Rev. Biochem. 70, 369–413 (2001)
Corbett, K. D. & Berger, J. M. Structure, molecular mechanisms, and evolutionary relationships in DNA topoisomerases. Annu. Rev. Biophys. Biomol. Struct. 33, 95–118 (2004)
Wang, J. C. DNA topoisomerases. Annu. Rev. Biochem. 65, 635–692 (1996)
Liu, L. F. et al. Mechanism of action of camptothecin. Ann. NY Acad. Sci. 922, 1–10 (2000)
Pommier, Y., Pourquier, P., Fan, Y. & Strumberg, D. Mechanism of action of eukaryotic DNA topoisomerase I and drugs targeted to the enzyme. Biochim. Biophys. Acta 1400, 83–105 (1998)
Sekiguchi, J. & Shuman, S. Vaccinia topoisomerase binds circumferentially to DNA. J. Biol. Chem. 269, 31731–31734 (1994)
Redinbo, M. R., Stewart, L., Kuhn, P., Champoux, J. J. & Hol, W. G. Crystal structures of human topoisomerase I in covalent and noncovalent complexes with DNA. Science 279, 1504–1513 (1998)
Cheng, C., Kussie, P., Pavletich, N. & Shuman, S. Conservation of structure and mechanism between eukaryotic topoisomerase I and site-specific recombinases. Cell 92, 841–850 (1998)
Kim, R. A. & Wang, J. C. Function of DNA topoisomerases as replication swivels in Saccharomyces cerevisiae . J. Mol. Biol. 208, 257–267 (1989)
Stewart, L., Redinbo, M. R., Qiu, X., Hol, W. G. & Champoux, J. J. A model for the mechanism of human topoisomerase I. Science 279, 1534–1541 (1998)
Stivers, J. T., Harris, T. K. & Mildvan, A. S. Vaccinia DNA topoisomerase I: evidence supporting a free rotation mechanism for DNA supercoil relaxation. Biochemistry 36, 5212–5222 (1997)
Brown, P. O. & Cozzarelli, N. R. A sign inversion mechanism for enzymatic supercoiling of DNA. Science 206, 1081–1083 (1979)
Brown, P. O. & Cozzarelli, N. R. Catenation and knotting of duplex DNA by type 1 topoisomerases: a mechanistic parallel with type 2 topoisomerases. Proc. Natl Acad. Sci. USA 78, 843–847 (1981)
Brown, P. O., Peebles, C. L. & Cozzarelli, N. R. A topoisomerase from Escherichia coli related to DNA gyrase. Proc. Natl Acad. Sci. USA 76, 6110–6114 (1979)
Liu, L. F., Liu, C. C. & Alberts, B. M. Type II DNA topoisomerases: enzymes that can unknot a topologically knotted DNA molecule via a reversible double-strand break. Cell 19, 697–707 (1980)
Mizuuchi, K., Fisher, L. M., O'Dea, M. H. & Gellert, M. DNA gyrase action involves the introduction of transient double-strand breaks into DNA. Proc. Natl Acad. Sci. USA 77, 1847–1851 (1980)
Dekker, N. H. et al. The mechanism of type IA topoisomerases. Proc. Natl Acad. Sci. USA 99, 12126–12131 (2002)
Strick, T. R., Croquette, V. & Bensimon, D. Single-molecule analysis of DNA uncoiling by a type II topoisomerase. Nature 404, 901–904 (2000)
Strick, T. R., Allemand, J. F., Bensimon, D., Bensimon, A. & Croquette, V. The elasticity of a single supercoiled DNA molecule. Science 271, 1835–1837 (1996)
Rice, J. A. Mathematical Statistics and Data Analysis 31–59 (Wadsworth & Brooks/Cole Advanced Books & Software, Pacific Grove, 1988)
Shuman, S., Bear, D. G. & Sekiguchi, J. Intramolecular synapsis of duplex DNA by vaccinia topoisomerase. EMBO J. 16, 6584–6589 (1997)
Sekiguchi, J. & Shuman, S. Requirements for noncovalent binding of vaccinia topoisomerase I to duplex DNA. Nucleic Acids Res. 22, 5360–5365 (1994)
Sekiguchi, J. & Shuman, S. Identification of contacts between topoisomerase I and its target DNA by site-specific photocrosslinking. EMBO J. 15, 3448–3457 (1996)
Carey, J. F., Schultz, S. J., Sisson, L., Fazzio, T. G. & Champoux, J. J. DNA relaxation by human topoisomerase I occurs in the closed clamp conformation of the protein. Proc. Natl Acad. Sci. USA 100, 5640–5645 (2003)
Woo, M. H. et al. Locking the DNA topoisomerase I protein clamp inhibits DNA rotation and induces cell lethality. Proc. Natl Acad. Sci. USA 100, 13767–13772 (2003)
Moroz, J. D. & Nelson, P. Torsional directed walks, entropic elasticity, and DNA twist stiffness. Proc. Natl Acad. Sci. USA 94, 14418–14422 (1997)
Bustamante, C., Bryant, Z. & Smith, S. B. Ten years of tension: single-molecule DNA mechanics. Nature 421, 423–427 (2003)
Krogh, B. O. & Shuman, S. DNA strand transfer catalyzed by vaccinia topoisomerase: peroxidolysis and hydroxylaminolysis of the covalent protein-DNA intermediate. Biochemistry 39, 6422–6432 (2000)
Shuman, S., Golder, M. & Moss, B. Characterization of vaccinia virus DNA topoisomerase I expressed in Escherichia coli . J. Biol. Chem. 263, 16401–16407 (1988)
Acknowledgements
We thank P. Veenhuizen for help in constructing various DNA constructs, D. Bensimon, U. Keyser, R. Seidel, K. Neuman, L. Tian and B. Spanjaard for stimulating discussions, C. Wiggins for advice on statistical data analysis, D. Lubensky for discussions on polymer physics, and J. van der Does for machining of flow cells. We thank FOM and NWO for financial support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Rights and permissions
About this article
Cite this article
Koster, D., Croquette, V., Dekker, C. et al. Friction and torque govern the relaxation of DNA supercoils by eukaryotic topoisomerase IB. Nature 434, 671–674 (2005). https://doi.org/10.1038/nature03395
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature03395
This article is cited by
-
Supercoiled DNA Minicircles under Double-strand Breaks
Chinese Journal of Polymer Science (2024)
-
CRISPR–Cas12a-mediated DNA clamping triggers target-strand cleavage
Nature Chemical Biology (2022)
-
Scanning probe microscopy
Nature Reviews Methods Primers (2021)
-
The RNA polymerase I transcription inhibitor CX-5461 cooperates with topoisomerase 1 inhibition by enhancing the DNA damage response in homologous recombination-proficient high-grade serous ovarian cancer
British Journal of Cancer (2021)
-
The free energy landscape of retroviral integration
Nature Communications (2019)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.