Abstract
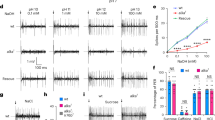
The sense of taste provides animals with valuable information about the nature and quality of food. Bitter taste detection functions as an important sensory input to warn against the ingestion of toxic and noxious substances. T2Rs are a family of approximately 30 highly divergent G-protein-coupled receptors (GPCRs)1,2 that are selectively expressed in the tongue and palate epithelium1 and are implicated in bitter taste sensing1,2,3,4,5,6,7,8. Here we demonstrate, using a combination of genetic, behavioural and physiological studies, that T2R receptors are necessary and sufficient for the detection and perception of bitter compounds, and show that differences in T2Rs between species (human and mouse) can determine the selectivity of bitter taste responses. In addition, we show that mice engineered to express a bitter taste receptor in ‘sweet cells’9 become strongly attracted to its cognate bitter tastants, whereas expression of the same receptor (or even a novel GPCR) in T2R-expressing cells resulted in mice that are averse to the respective compounds. Together these results illustrate the fundamental principle of bitter taste coding at the periphery: dedicated cells act as broadly tuned bitter sensors that are wired to mediate behavioural aversion.
This is a preview of subscription content, access via your institution
Access options
Subscribe to this journal
Receive 51 print issues and online access
$199.00 per year
only $3.90 per issue
Buy this article
- Purchase on Springer Link
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
References
Adler, E. et al. A novel family of mammalian taste receptors. Cell 100, 693–702 (2000)
Matsunami, H., Montmayeur, J. P. & Buck, L. B. A family of candidate taste receptors in human and mouse. Nature 404, 601–604 (2000)
Chandrashekar, J. et al. T2Rs function as bitter taste receptors. Cell 100, 703–711 (2000)
Bufe, B., Hofmann, T., Krautwurst, D., Raguse, J. D. & Meyerhof, W. The human TAS2R16 receptor mediates bitter taste in response to beta-glucopyranosides. Nature Genet. 32, 397–401 (2002)
Kim, U. K. et al. Positional cloning of the human quantitative trait locus underlying taste sensitivity to phenylthiocarbamide. Science 299, 1221–1225 (2003)
Behrens, M. et al. The human taste receptor hTAS2R14 responds to a variety of different bitter compounds. Biochem. Biophys. Res. Commun. 319, 479–485 (2004)
Pronin, A. N., Tang, H., Connor, J. & Keung, W. Identification of ligands for two human bitter T2R receptors. Chem. Senses 29, 583–593 (2004)
Kuhn, C. et al. Bitter taste receptors for saccharin and acesulfame K. J. Neurosci. 24, 10260–10265 (2004)
Zhao, G. Q. et al. The receptors for mammalian sweet and umami taste. Cell 115, 255–266 (2003)
Zhang, Y. et al. Coding of sweet, bitter, and umami tastes: different receptor cells sharing similar signaling pathways. Cell 112, 293–301 (2003)
Caicedo, A. & Roper, S. D. Taste receptor cells that discriminate between bitter stimuli. Science 291, 1557–1560 (2001)
Farbman, A. I. Electron microscope study of the developing taste bud in rat fungiform papilla. Dev. Biol. 11, 110–135 (1965)
Coward, P. et al. Controlling signaling with a specifically designed Gi-coupled receptor. Proc. Natl Acad. Sci. USA 95, 352–357 (1998)
Glendinning, J. I., Gresack, J. & Spector, A. C. A high-throughput screening procedure for identifying mice with aberrant taste and oromotor function. Chem. Senses 27, 461–474 (2002)
McBride, K., Slotnick, B. & Margolis, F. L. Does intranasal application of zinc sulfate produce anosmia in the mouse? An olfactometric and anatomical study. Chem. Senses 28, 659–670 (2003)
Franchini, L. F., Rubinstein, M. & Vivas, L. Reduced sodium appetite and increased oxytocin gene expression in mutant mice lacking beta-endorphin. Neuroscience 121, 875–881 (2003)
Nelson, G. et al. Mammalian sweet taste receptors. Cell 106, 381–390 (2001)
Dahl, M., Erickson, R. P. & Simon, S. A. Neural responses to bitter compounds in rats. Brain Res. 756, 22–34 (1997)
Nelson, G. et al. An amino-acid taste receptor. Nature 416, 199–202 (2002)
Danilova, V. & Hellekant, G. Comparison of the responses of the chorda tympani and glossopharyngeal nerves to taste stimuli in C57BL/6J mice. BMC Neurosci. 4, 5 (2003)
Nakashima, K. & Ninomiya, Y. Transduction for sweet taste of saccharin may involve both inositol 1,4,5-trisphosphate and cAMP pathways in the fungiform taste buds in C57BL mice. Cell. Physiol. Biochem. 9, 90–98 (1999)
Acknowledgements
We thank F. Liu for help generating T2R5 knockout animals. We are also grateful to A. Cho and S. Taduru for generation of transgenic lines, and to A. Leslie for preparation of antibodies. We thank members of the Zuker and Ryba laboratories for valuable comments and advice. This work was supported in part by a grant from the National Institute on Deafness and Other Communication Disorders to C.S.Z. C.S.Z. is an investigator of the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Competing interests
The authors declare that they have no competing financial interests.
Supplementary information
Supplementary Figure Legends
This file contains the figure legends for Supplementary Figures 1-3. (DOC 22 kb)
Supplementary Figure 1
T2R5-/- construct and physiology. (PDF 84 kb)
Supplementary Figure 2
T2R5-/- mice respond normally to a variety of bitter, sweet, umami, salty and sour stimuli. (PDF 56 kb)
Supplementary Figure 3
T2R cells mediate bitter but not sweet or umami taste. (PDF 77 kb)
Rights and permissions
About this article
Cite this article
Mueller, K., Hoon, M., Erlenbach, I. et al. The receptors and coding logic for bitter taste. Nature 434, 225–229 (2005). https://doi.org/10.1038/nature03352
Received:
Accepted:
Issue Date:
DOI: https://doi.org/10.1038/nature03352
This article is cited by
-
Bitter taste receptor activation by cholesterol and an intracellular tastant
Nature (2024)
-
Polycystic kidney disease 2-like 1 channel contributes to the bitter aftertaste perception of quinine
Scientific Reports (2023)
-
An update on extra-oral bitter taste receptors
Journal of Translational Medicine (2021)
-
Expression of Eya1 in mouse taste buds
Cell and Tissue Research (2021)
Comments
By submitting a comment you agree to abide by our Terms and Community Guidelines. If you find something abusive or that does not comply with our terms or guidelines please flag it as inappropriate.